Abstract 摘要
Two decades of research on RNA interference (RNAi) have transformed a breakthrough discovery in biology into a robust platform for a new class of medicines that modulate mRNA expression. Here we provide an overview of the trajectory of small-interfering RNA (siRNA) drug development, including the first approval in 2018 of a liver-targeted siRNA interference (RNAi) therapeutic in lipid nanoparticles and subsequent approvals of five more RNAi drugs, which used metabolically stable siRNAs combined with N-acetylgalactosamine ligands for conjugate-based liver delivery. We also consider the remaining challenges in the field, such as delivery to muscle, brain and other extrahepatic organs. Today’s RNAi therapeutics exhibit high specificity, potency and durability, and are transitioning from applications in rare diseases to widespread, chronic conditions.
二十年的 RNA 干扰(RNAi)研究将生物学中的突破性发现转变为一种强大的平台,用于调节 mRNA 表达的新型药物。在这里,我们概述了小干扰 RNA(siRNA)药物开发的历程,包括 2018 年首个针对肝脏的 siRNA 干扰(RNAi)治疗药物在脂质纳米颗粒中的批准,以及随后五种 RNAi 药物的批准,这些药物使用了代谢稳定的 siRNA 与 N-乙酰半乳糖胺配体结合进行肝脏递送。我们还考虑了该领域剩余的挑战,例如向肌肉、大脑和其他肝外器官的递送。如今的 RNAi 治疗药物展现出高特异性、效力和持久性,并正从罕见疾病的应用转向广泛的慢性病。
Similar content being viewed by others
其他人正在查看的类似内容
Main 主
The journey of RNA interference (RNAi) for therapeutic applications began with the seminal discovery of the RNAi mechanism in 1998 by Fire and Mello1, who showed that the expression of specific genes in Caenorhabditis elegans could be silenced using long double-stranded RNA (dsRNA). Building on this, work in Tuschl’s2 laboratory, carried out in 2001, demonstrated that sequence-specific RNAi can be achieved in mammalian cells using small-interfering RNAs (siRNAs), without inducing the immune response seen with long dsRNA2. These discoveries put RNAi on the radar of drug researchers looking for potent ways to regulate gene expression, and led to efforts to harness RNAi for therapeutic purposes.
RNA 干扰(RNAi)在治疗应用中的旅程始于 1998 年 Fire 和 Mello 的开创性发现,他们展示了在秀丽隐杆线虫中可以使用长双链 RNA(dsRNA)沉默特定基因的表达。在此基础上,Tuschl 实验室于 2001 年的研究表明,可以使用小干扰 RNA(siRNA)在哺乳动物细胞中实现序列特异性的 RNAi,而不会引发长 dsRNA 所见的免疫反应。这些发现使 RNAi 引起了药物研究人员的关注,他们寻找有效的方式来调控基因表达,并推动了利用 RNAi 进行治疗的努力。
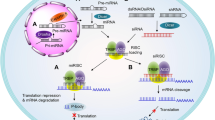
Therapeutics based on siRNA and antisense oligonucleotides (ASOs) are related to, but distinct from, endogenous non-coding RNAs, such as microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and piwi-interacting RNAs (piRNAs)3,4,5,6, that regulate gene expression at the epigenetic, transcriptional or translational level. Exogenous ASOs and siRNAs typically enter cells through endocytosis and require extensive chemical modifications or protection through a supramolecular delivery system to confer the stability required to escape a metabolically highly active endo-lysosomal compartment7. ASOs, the first oligonucleotide drugs to gain regulatory approval, modulate pre-messenger RNA (mRNA) splicing8 (Fig. 1a) or engage endogenous ribonuclease H after recognition of target RNA7 (Fig. 1b). By contrast, siRNAs harness the endogenous RNA-induced silencing complex (RISC)9, a multi-protein system consisting of Argonaute 2 (AGO2), trans-activation response RNA binding protein 2 and DICER1 (Fig. 1c). Unlike single-stranded ASOs, siRNAs are short dsRNAs, comprising a ‘passenger’ (sense) strand that has the same sequence as the target RNA and a complementary ‘guide’ (antisense (AS)) strand10,11. This structure facilitates the interaction of siRNAs and miRNAs, the natural counterparts of synthetic siRNAs, with RISC12. During this process, the passenger strand is removed, allowing the guide strand to serve as a template for target recognition11. Most naturally occurring miRNAs bind to their targets with low affinity through partial sequence match, resulting in destabilization or translational inhibition of target mRNA13,14 (Fig. 1d). By contrast, most synthetic siRNAs are designed to be fully complementary to their target site, resulting in high-affinity binding and RISC-mediated cleavage of the target transcript through AGO2 endonuclease12,15. In this multi-turnover catalytic process, a single RISC-loaded siRNA molecule, with an estimated half-life of 4 to 5 d in rodents, can degrade thousands of target RNA molecules16,17. The combination of exceptional specificity, potency and durability of effect endows siRNA with the clinically desirable property of requiring very infrequent dosing18.
基于 siRNA 和反义寡核苷酸(ASOs)的治疗与内源性非编码 RNA(如微小 RNA(miRNAs)、长非编码 RNA(lncRNAs)和 piwi 相互作用 RNA(piRNAs))相关,但又有所不同,这些内源性 RNA 在表观遗传、转录或翻译水平上调节基因表达。外源性 ASOs 和 siRNAs 通常通过内吞作用进入细胞,并需要广泛的化学修饰或通过超分子递送系统进行保护,以赋予其逃避代谢高度活跃的内溶酶体区室所需的稳定性。ASOs 是首批获得监管批准的寡核苷酸药物,调节前信使 RNA(mRNA)剪接或在识别目标 RNA 后与内源性核糖核酸酶 H 结合。相比之下,siRNAs 利用内源性 RNA 诱导沉默复合体(RISC),这是一个由 Argonaute 2(AGO2)、转激活反应 RNA 结合蛋白 2 和 DICER1 组成的多蛋白系统。与单链 ASOs 不同,siRNAs 是短双链 RNA,由与目标 RNA 具有相同序列的“乘客”(正义)链和互补的“引导”(反义(AS))链组成。 这种结构促进了 siRNA 和 miRNA(合成 siRNA 的天然对应物)与 RISC 的相互作用。在此过程中,乘客链被去除,使得引导链可以作为靶标识别的模板。大多数自然存在的 miRNA 通过部分序列匹配以低亲和力结合其靶标,导致靶 mRNA 的不稳定或翻译抑制。相比之下,大多数合成 siRNA 被设计为与其靶位点完全互补,从而导致高亲和力结合,并通过 AGO2 内切酶介导靶转录本的切割。在这个多轮催化过程中,单个 RISC 加载的 siRNA 分子在啮齿动物中估计的半衰期为 4 到 5 天,可以降解数千个靶 RNA 分子。卓越的特异性、效力和持久性使 siRNA 具备了临床上所需的非常少量给药的特性。
a, Synthetic (exogenous) ASOs can be designed to target splicing signal sequences and modulate pre-mRNA splicing7,9,137. b, ASOs can also be designed to bind complementary sequences in target mRNAs and recruit ribonuclease H1 (RNase H1) for ASO–RNA heteroduplex-mediated degradation of target mRNA9,138. c, Exogenous siRNAs are the cousins of endogenous miRNAs and utilize siRNA-induced silencing complex (siRISC) to mediate gene silencing, as described in the text. d, miRNAs utilize miRNA-induced silencing complex (miRISC) to mediate gene silencing through mRNA de-capping and poly(A)-tail shortening139,140,141.
Six siRNA therapeutics have already been approved by the US Food and Drug Administration (FDA), validating the RNAi approach for use in humans. This success was built on years of painstaking technological development and insights from several setbacks. We begin this Perspective by describing the early challenges and milestones in the field, including the formidable task of delivering exogenous siRNA to its intracellular site of action. We evaluate key considerations in designing siRNAs and selecting targets, describe preclinical evaluation of safety and tolerability and provide an overview of approved RNAi therapeutics and those that have advanced into late-stage clinical trials. Finally, we discuss expansion into extrahepatic targets and common diseases, and new areas where we expect RNAi to make an impact as an integral part of the new era of nucleic acid therapeutics.
Platform technologies for RNAi therapeutics
The creation of RNAi therapeutics was dependent on several advances in siRNA chemistry, as well as the development of complementary delivery technologies. In this section, we discuss the chemistry of early designs, the evolution of lipid nanoparticle (LNP) technology and the emergence of advanced chemistry designs and conjugates.
Early molecules
In our view, key reasons for the protracted period between the discovery of RNAi and the regulatory approval of the first RNAi therapeutic (Fig. 2) were the deficient drug-like properties of the early RNAi molecules and the lack of suitable systems for safe and efficient delivery to tissues of interest. Naked, unmodified RNA and DNA molecules are rapidly degraded in the body by endogenous nucleases and can be recognized by the immune system19,20. Furthermore, in early attempts to chemically modify siRNAs, it was challenging to achieve high metabolic stability without compromising RNAi activity.
The initial period of development between 2002 and 2008 was one of hope and promise, marked by early technological advances21. Several candidate siRNAs entered clinical development21,22, and the Nobel Prize in Physiology or Medicine was awarded to Fire and Mello in 2006. However, the following years saw many setbacks and the failures of several clinical programs. In retrospect, suboptimal siRNA designs and/or delivery systems with inadequate safety and efficacy profiles could not support late-stage clinical development. Examples include bevasiranib (Cand5) for wet, age-related macular degeneration in 2009 (ref. 23) and ALN-RSV for the treatment of respiratory syncytial virus, both of which were locally administered, essentially unmodified siRNAs without any features to improve delivery. Similarly, none of the programs that combined systemically administered siRNAs with early-generation delivery systems advanced into late-stage clinical development. Examples include ALN-VSP02 for solid tumors with liver involvement, and ALN-TTR01 for the treatment of hereditary transthyretin-mediated (hATTR) amyloidosis. In both cases, partially modified siRNAs were formulated in an early generation of LNPs for delivery (see below).
Consequently, early enthusiasm for RNAi gave way to doubts, and several big pharmaceutical companies exited the field between 2009 and 2013 (ref. 24). Ironically, during the same period, major breakthroughs were being made in the underlying siRNA chemistry and delivery technologies, which ultimately paved the way to successful translation in clinical studies.
LNP technology
LNPs are multicomponent lipid formulations that were originally developed to deliver DNA plasmids and ASOs. Key components of these formulations are ionizable lipids. These mediate encapsulation of nucleic acids and self-assembly of the particles during formulation, and promote endosomal release of the nucleic acids once the LNP is taken up by a cell18,25,26,27.
The first demonstration of RNAi activity in a higher species was achieved in 2006 using an early generation of LNPs called stable nucleic acid–lipid particles28. These included the ionizable lipid 1,2-dilinoleyloxy-N,N-dimethyl-3-aminopropane (DLinDMA), 3-N-[(methoxypoly(ethylene glycol)2000)carbamoyl]-1,2-dimyristyloxy-propylamine (PEG-C-DMA), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) and cholesterol28. In addition, the selective incorporation of 2′-O-methyl (2′-OMe) uridine or guanosine nucleosides was essential for abrogating the immunostimulatory activity of siRNAs29. Although these early results showed promise for LNP-based systemic delivery of RNAi therapeutics, they also indicated that the technology had potential limitations, due to its rather modest activity and narrow therapeutic window. Consequently, we and others in the field focused our efforts on identifying new lipids and formulations to improve nucleic acid delivery30. In collaboration with the Langer and Anderson laboratories at the Massachusetts Institute of Technology and the Cullis laboratory at the University of British Columbia, as well as AlCana Technologies and Tekmira Pharmaceuticals, we embarked on a multi-year lipid-discovery effort, during which thousands of lipids and formulations were designed and evaluated30,31,32.
In this process, we discovered that intravenously administered LNPs containing ionizable lipids can be targeted to hepatocytes by recruitment and binding of endogenous apolipoprotein E, which promotes hepatocellular uptake through receptor-mediated endocytosis33. This effort eventually led to the discovery of (6Z,9Z,28Z,31Z)-heptatriaconta-6,9,28,31-tetraen-19-yl-4-(dimethylamino)butanoate (DLin-MC3-DMA, also known as MC3), an ionizable lipid that improved the potency of siRNAs delivered by LNPs by approximately 100-fold over that of previous LNP formulations (Fig. 3a)34,35.
These results, together with an acceptable preclinical safety profile, led Alnylam Pharmaceuticals to advance into clinical development ALN-PCS02 and ALN-TTR02, two investigational RNAi therapies based on partially modified siRNAs (endo-light design; see Fig. 4a) formulated in MC3-containing second-generation LNPs36,37. The early clinical results of ALN-TTR02 demonstrated robust translation in humans and improved activity compared with ALN-TTR01, which consisted of the same siRNA molecule but used the earlier LNP formulation based on stable nucleic acid–lipid particles. ALN-TTR02, now known as patisiran, targets the liver-produced protein transthyretin (TTR) for the treatment of hATTR amyloidosis with polyneuropathy. In 2018, it became the first approved RNAi therapeutic and the first nucleic acid therapeutic delivered by LNPs38.
Individual structures are shown; modification patterns may vary depending on the specific compound; all structures are drawn with the SS at the top (5′ to 3′) and the AS at the bottom. a, Conventional, partially modified symmetrical design that features 2-nt DNA overhangs and a sequence-specific modification pattern (endo-light), with 2′-OMe modifications applied to all pyrimidines in the SS and to pyrimidines that are 5′ adjacent to the ribo A nucleoside. b, Fully modified 21- to 23-nt siRNAs with 2 PS linkages and a single 2-nt overhang (STC). c, Fully modified 21- to 23-nt siRNAs with 6 PS linkages and a single 2-nt overhang (ESC). d, Fully modified 21- to 23-base siRNAs with 6 PS linkages, a single 2-nt overhang and thermally destabilizing modification (for example, GNA) in the seed region (ESC+). e, Fully modified 19- to 21-base duplex with various end modifications but no overhangs (targeted RNAi molecule (TRiM)). f, Fully modified, nicked galanin (GalXC) Dicer-substrate design comprising a 24- to 26-nt duplex with a hairpin loop structure carrying targeting ligands. g, Fully modified short asymmetric siRNA design with long PS-stabilized AS overhang (self-delivering siRNAs (sd-rxRNAs)).
Efforts to identify lipids and LNP formulations with even wider therapeutic indexes are ongoing, in part driven by the need to deliver larger mRNA payloads for therapeutic applications and vaccines39, including mRNA vaccines against SARS-CoV-2 (ref. 40). Newer generations of LNPs incorporate biodegradable lipids containing ester linkages in the lipid tails, which are eliminated more rapidly and exhibit improved safety profiles in preclinical models compared with non-biodegradable lipids (Fig. 3b)41,42,43.
Conjugates and evolution of siRNA chemistry
In parallel with the work on LNP-based systems, our group at Alnylam Pharmaceuticals was advancing a conjugate-based, targeted delivery platform with the aim of creating a fully characterizable, single-component entity that could potentially be administered subcutaneously. The key elements that made this strategy successful were: (1) a high-capacity cell surface receptor on the cell type of interest, (2) a targeting ligand capable of binding with high affinity and specificity and (3) a nuclease-resistant siRNA. Earlier work had demonstrated that one can design fully chemically modified siRNAs that maintain their intrinsic potency (that is, their ability to interact with RISC) while exhibiting improved metabolic stability44,45. However, the breakthrough for siRNA conjugates was achieved several years later, when we covalently conjugated a trivalent N-acetylgalactosamine (GalNAc) ligand, targeting the asialoglycoprotein receptor (ASGPR) expressed on hepatocytes, to fully chemically modified siRNAs with additional terminal phosphorothioate (PS) linkages for enhanced metabolic stability46,47.
The choice of ASGPR as a target receptor was based on its high expression in hepatocytes48,49, rapid recycling time, ability to constitutively endocytose a multitude of large cargos and well-characterized preference for multivalent galactose or GalNAc-presenting ligands48,49,50,51. In addition, the receptor had already been used to bind GalNAc-terminated glycolipids and thereby to clear lipoprotein particles from circulation52. Consequently, Alnylam Pharmaceuticals and others reengineered well-characterized bi- and triantennary GalNAc ligands to facilitate covalent conjugation to siRNAs either during solid-phase oligonucleotide synthesis or through post-synthetic coupling53,54,55.
In 2013, revusiran became the first GalNAc–siRNA conjugate to enter clinical development56. Like patisiran, revusiran targeted TTR mRNA expressed in the liver; however, revusiran was evaluated as a treatment for hATTR amyloidosis with cardiomyopathy, a disease that is predominantly diagnosed in older people with heart failure. Despite its fully chemically modified structure, revusiran lacked efficient 5′ nuclease protection and had to be administered at high doses, initially in a daily loading phase (500 mg daily for 5 d) with weekly 500 mg doses thereafter. A phase 1 study in healthy volunteers demonstrated robust and dose-dependent reduction in circulating TTR protein with encouraging safety56. Unfortunately, an imbalance in deaths between the placebo and treatment groups during the phase 3 study led to the discontinuation in 2016 of the clinical development of this early-generation GalNAc conjugate. A careful analysis of the clinical data indicated that, at baseline, a greater percentage of participants who died while on treatment were older and showed evidence of more advanced heart failure compared with those who remained alive57. Nevertheless, the investigation could not establish a causal mechanism for the mortality imbalance, and therefore a role for revusiran and/or its metabolites, such as 2′-fluoronucleotides, cannot fully be excluded.
The discontinuation of the first GalNAc–siRNA conjugate program during late-stage clinical development represented a substantial setback for RNAi technology, with concerns about potential class effects affecting other clinical development programs using the same conjugate-based delivery approach. Fortunately, the conjugate-based approach continued to advance rapidly, thanks to the improved understanding of the intracellular compartmentalization of siRNAs and the crucial role of intracellular metabolic stability17,58. Guided by more sensitive bioanalytical assays and tools, such as liquid chromatography–mass spectrometry and intracellular imaging technologies, we found that GalNAc–siRNA conjugates remain trapped in endo-lysosomal compartments, which harbor different and more aggressive nuclease activities than those found in plasma. We rationalized that, by further improving the stability of the siRNAs against nucleolytic degradation, it might be possible to extend the window of time during which a siRNA can escape from this intracellular depot17 and enhance its potency and duration of effect. Indeed, the evolution of the siRNA design over the past 16 years highlights the importance of metabolic stability, and in vivo potency was improved by three orders of magnitude compared with the parent, largely unmodified molecules through judiciously placed chemical modifications that preserve the ability of the siRNA to engage with RISC59.
The continual advancements in siRNA design are also reflected in the succession of clinical development candidates. Whereas revusiran used the standard template chemistry (STC) siRNA design (Fig. 4b), subsequent candidates feature an enhanced stabilization chemistry (ESC) design that incorporates additional terminal PS modifications46. The additional stabilization of the 5′ ends of both strands addressed a major metabolic liability caused by the dominant 5′-exonuclease activity present in the endo-lysosomal compartment. Further improvements in siRNA design in the subsequent years focused on optimizing the 2′-fluoro (2′-F)/2′-OMe modification pattern. Our group at Alnylam Pharmaceuticals evaluated new designs across multiple sequences in parallel, on the basis of the statistical analysis of a large in vitro dataset combined with screening. The advanced ESC designs feature a more than 50% reduction in 2′-F content and a concomitant increase in 2′-OMe content across both strands of the siRNA (Fig. 4c)59. This approach provided additional protection against nuclease degradation without compromising intrinsic activity, yielding RNAi therapeutics that can be administered to people at much lower doses and at a frequency of monthly to biannually60,61. The tremendous impact of siRNA chemistry and metabolic stability becomes apparent in the comparison of revusiran and vutrisiran, two siRNAs with the same sequence but different chemical designs. Whereas revusiran required a staggering 28,000 mg cumulative annual dose, the follow-on molecule vutrisiran requires a 280-fold lower annual dose of 100 mg (given quarterly at 25 mg).
More recently, Alnylam Pharmaceuticals has introduced additional chemical modifications, such as glycol nucleic acid (GNA), to further improve the specificity of siRNAs in cases where the siRNAs have seed-mediated RNAi off-target effects62 (Fig. 4d, also see ‘Safety and tolerability’). Furthermore, stable phosphate mimics, such as 5′-(E)-vinylphosphonate, have been incorporated to enhance RISC loading63,64. This modification has been found particularly useful for certain extrahepatic tissues, such as those in the central nervous system (CNS)65, in which the intrinsic kinase activity may be too low for the efficient 5′ phosphorylation that is required for stable RISC loading.
Discovery and preclinical development
The selection of therapeutic targets has generally focused on genes validated by human genetics, the presence of a well-established circulating biomarker to assess activity and a clear development path toward registration. These elements minimize target risk and allow direct assessment of clinical translation.
siRNA lead discovery
The lead discovery process, from the initial design through preclinical proof of concept to the selection of the candidate for further preclinical development, generally takes 9–12 months. The process starts with an in silico analysis of all possible siRNAs against the intended target transcript, using prediction algorithms for potency and specificity. A subset of siRNAs, typically a few hundred compounds, is then synthesized using a set of established chemical-modification patterns and templates and screened for on- and off-target activity in cell culture, followed by in vivo evaluation of pharmacodynamics (PD) in rodents. A small number of compounds that emerge from this optimization process, which includes non-clinical safety studies, advance into on-target pharmacology studies in non-human primates66,67. Not all target RNAs are equally amenable to RNAi. Features like high turnover of mRNA are not favorable for effective knockdown by siRNAs68. The clinical doses also depend on the extent of knockdown required for therapeutic benefit. For example, an approximately 80% knockdown of serum TTR protein is sufficient for treatment of polyneuropathy, whereas complement factor 5 (C5) must be reduced by >99% for the successful treatment of paroxysmal nocturnal hemoglobinuria.
Reproducibility and predictability of pharmacokinetics and pharmacodynamics across species
Following subcutaneous administration, GalNAc-conjugated siRNAs are rapidly absorbed into the systemic circulation and distributed predominantly to the liver hepatocytes69. Direct renal excretion is a minor elimination pathway for GalNAc-conjugated siRNAs69. This is likely because subcutaneous dosing generally allows the levels of GalNAc–siRNA entering the circulation to remain below the ASGPR saturation point, allowing efficient uptake in hepatocytes. Extensive preclinical characterization of numerous GalNAc-conjugated siRNAs demonstrates that their absorption, distribution, metabolism, excretion and PK and PD properties are generally well-conserved across species67. This enables the prediction of human PK and PD and translation of the clinical dose and dosing regimen. Even across different compounds, the overall dose-normalized exposures are generally within twofold of the mean for plasma and liver, resulting in highly predictable and extrapolatable plasma and liver PK properties.
Unlike traditional small-molecule drugs, RNAi therapeutics are a truly modular platform. The pharmacophore is largely contained in the siRNA sequence, allowing the application of chemistry insights across molecules, even across compounds using different delivery ligands. The design, which encompasses siRNA strand length, the chemical-modification pattern and the nature of the delivery system, can be tailored independently of the siRNA sequence to achieve the desired target product profile, such as potency, duration of effect, specificity and biodistribution70. Thus, as mentioned above, a given delivery system, such as a GalNAc ligand, applied to different siRNAs will result in similar PK and biodistribution profiles67,70,71.
Existing chemical-modification patterns limit metabolic degradation. This phenomenon seems to be well-conserved across species, with rodents exhibiting an overall higher nuclease activity than non-human primates and humans67. With only a small portion of GalNAc-conjugated siRNAs (typically <10%) being metabolized in plasma, the majority is metabolized slowly in the liver and excreted as metabolites through urine and bile67. The low rate of degradation results in an intracellular depot17 supporting an extended duration of effect that is suitable for infrequent dosing in the clinic.
Safety and tolerability
Potential safety liabilities of RNAi therapeutics were assessed in three main categories: toxicities associated with the intracellular accumulation of siRNAs and their metabolites, perturbation of endogenous RNAi pathways and hybridization-based off-target effects. Regarding the potential chemical toxicities associated with the siRNAs or their metabolites, particular focus has been placed on the non-natural modifications, such as 2′-F and PS, used to stabilize the siRNA molecules.
The 2′-fluororibo modification is an excellent mimic of natural RNA, as well as DNA, and increases resistance to nuclease degradation72. 2′-fluororibo pyrimidines (2′-F-uridine and cytidine) were first used in macugen, an aptamer-based drug approved for the treatment of age-related macular degeneration73. Because fialuridine—a nucleoside analog carrying 2′-F in the arabino (up) configuration, as well as 5-iodo on the uracil base—had been associated with severe liver toxicity in clinical studies74, the 2′-fluororibo pyrimidines underwent an extensive safety evaluation in preclinical studies before human use75. Neither compound showed evidence of the toxicity in rat or woodchucks comparable to that observed with fialuridine, and none of the observed effects were considered adverse. Nevertheless, it has been shown that 2′-F nucleotides can be incorporated into DNA and RNA in vitro and in vivo, albeit at high doses75,76. Furthermore, the 2′-F modification has been implicated in causing in vitro cytotoxicity when used in single-stranded ASOs with full PS backbone modifications77. Because all four 2′-fluororibo nucleosides (U, C, A and G) were incorporated into the first GalNAc–siRNA conjugate, revusiran56, we carried out a comprehensive safety investigation including all four 2′-F nucleotides. We found that 2′-F-nucleoside triphosphates are weak inhibitors and poor substrates for mitochondrial DNA and RNA polymerases, do not act as chain terminators and are efficiently out-competed by native nucleotides in polymerase reactions78. Furthermore, in our preclinical and clinical studies, even with STC GalNAc–siRNAs dosed chronically at suprapharmacologic levels, we have not observed any evidence of liver failure or nephropathy. Causes of the observed mortality imbalance associated with revusiran were thoroughly investigated, but no clear causative mechanism was identified. Although the results suggested that cardiac parameters in both treatment arms progressed similarly, a role for revusiran and/or its metabolites, such as 2′-fluronucleosides, cannot fully be excluded57. Our extensive safety assessments affirm that the overall risk for mitochondrial toxicity or other toxic side effects mediated by 2′-F-monomer metabolites is low, and 2′-F modifications have subsequently been applied to essentially all conjugate-based RNAi therapeutics that are either already approved or currently in clinical development. Recently, a report on 4-year safety data for inclisiran indicates that chronic dosing with this molecule seems to be pharmacologically active and well tolerated79.
The other main non-natural chemical modification used in current siRNA conjugates is modification of the phosphate backbone with PS linkages to enhance metabolic stability80,81, an approach pioneered by Eckstein82 and first used in the ASO field83. Although extensive use of PS modifications has been associated with adverse findings84, these effects have not been seen with siRNAs85,86. This difference is likely owing to the small number of PS modifications that is required to stabilize siRNA, typically six to eight PS linkages spread across both strands, and the well-defined, double-stranded structure of siRNAs. This design predominantly exposes the hydrophilic sugar–phosphate backbone, thereby minimizing non-specific interactions.
In addition to the risk of toxicity originating from the chemically modified sugar–phosphate backbone, there is also the potential for competition of exogenous siRNAs with endogenous miRNAs for components of the RNAi pathway, thereby interfering with miRNA function. Indeed, this was observed for short hairpin RNAs that were expressed under a strong promoter, which saturated the exportin molecule that transports precursor miRNAs (pre-miRNAs) from the nucleus to the cytoplasm87,88. However, exogenous siRNAs bypass this pathway and engage directly with cytosolic RISC. This finding was further supported in mechanistic studies with hepatotoxic siRNAs using Reversir technology, which can specifically block RISC-loaded siRNA89. The Reversir treatment abrogated hepatotoxicity without affecting RISC loading, suggesting that hepatotoxicity was not caused by competition for RISC complexes with endogenous RNAi pathways62.
Lastly, RISC-loaded siRNAs could elicit RNAi-mechanism-based off-target effects in a miRNA-like fashion through seed-region-mediated interaction with off-target transcripts, resulting in mRNA destabilization and degradation90,91,92. A thorough mechanistic evaluation found that the hepatotoxicity observed for a subset of GalNAc–siRNA conjugates could largely be attributed to RNAi-mediated off-target effects through seed matches62. Although these off-target effects are harder to avoid because there can be many transcripts with short seed-based matches of ~7 nucleotides (nt), they can be mitigated by sequence selection and/or modulation of seed-mediated transcript binding strength through thermally destabilizing chemical modifications (ESC+ design)62,93,94. This concept was further validated in preclinical species and subsequently in humans, as GalNAc–siRNA conjugates that had been previously associated with liver enzyme elevations showed an improved safety profile following the incorporation of a single GNA in the seed region95,96. With no evidence of chemical toxicity and a low propensity for RNAi pathway perturbations, the siRNA designs currently used for the GalNAc conjugates offer the potential for exquisite specificity.
Clinical development
Although synthetic siRNAs generally have the double-stranded structure of endogenous miRNAs, a few designs have advanced into clinical development97 (Fig. 4). The ‘classical’ design-which, for instance, was applied to the LNP-formulated patisiran—features 2-nt overhangs on the 3′ ends of a 19-base-pair duplex2 (Fig. 4a). Attempts to reduce the size and to combine the double-stranded structure of siRNAs with some features of single-stranded ASOs have led to asymmetric siRNA designs, in which a 19- to 21-nt AS is paired with a shorter 11- to 15-nt sense strand (SS)98 (Fig. 4g). The long overhang of the AS is stabilized with PS linkages, which may impact extrahepatic tissue distribution and efficacy99. At the other end of the size spectrum are Dicer-substrate siRNAs, 25- to 30-base-pair duplexes that were originally designed to be recognized and converted into shorter 19- to 21-nt-long siRNAs by the endonuclease Dicer, an enzyme in the RNAi pathway that is responsible for pre-miRNA processing (Fig. 4f)100. With the broad adoption of the GalNAc conjugate strategy, the designs now generally include extensive use of 2′ modifications, such as 2′-OMe and 2′-F across the entire siRNA, combined with stabilization of the ends with PS and/or other chemical modifications. Most RNAi therapeutics that are currently approved or are in clinical development feature a 19- to 21-nt duplex structure with a single 2-nt overhang at the 3′ end of the AS strand (Fig. 4c). We found that this structure, at least in the context of fully modified GalNAc–siRNA conjugates, seems to be preferred by RISC (likely because of the inbuilt bias for RISC loading of the AS promoted by the single overhang structure) and shows excellent in vivo translation across species. Other structures, such as double 3′ overhangs on each SS and AS or blunt ends (no overhangs) can also work well, likely as a consequence of the metabolic stability provided by this design with exonuclease protection at both ends of each strand.
Approved therapies
In 2018, patisiran became the first approved RNAi therapeutic. Thus far, it remains the only approved LNP-formulated siRNA38. The other five approved RNAi drugs are GalNAc–siRNA conjugates, the most recent being nedosiran in 2023 (Table 1). All six current FDA-approved RNAi therapeutics target transcripts expressed in the liver38. Several additional liver-targeted development candidates are in late-stage trials97, and many more are in earlier stages of clinical development across different disease areas.
Therapies in late-stage trials
The success of GalNAc-conjugated siRNA technology has transformed the field of nucleic acid therapeutics. Although LNPs offer a faster onset of action101, the favorable and consistent preclinical and clinical activity of subcutaneously dosed GalNAc conjugates, combined with the less complex delivery system, have established them as the predominant liver-targeted system for oligonucleotide therapeutics, including ASOs71. Several investigational liver-directed RNAi therapeutics are in late-stage clinical trials. Notably, the advancement of multiple GalNAc conjugates, which use chemically fully modified siRNAs (Fig. 4c–f), through late-stage clinical development further validates the GalNAc conjugate approach and highlights its safe and effective use in humans across targets and indications.
Outlook
As described above, a range of RNAi therapeutics are now in use or development for the treatment of a wide variety of liver-based diseases70,97,102. The latest advances to the GalNAc conjugate platform include durable molecules that provide sustained pharmacology potentially suitable for annual dosing. This duration of action, unprecedented for a drug that does not alter the genome, comes with the pharmacologic advantages of RNAi therapeutics: a predictable, dose-responsive PK–PD profile and a reversible mechanism of action without permanent effects on the genome.
Expansion into prevalent diseases and combination therapies
The approval of inclisiran marked the extension of RNAi therapeutics into the space of common diseases and highlights the disruptive potential of long-acting drugs. Drugs that offer sustained reductions in risk factors, such as low-density lipoprotein-cholesterol (LDL-C), may provide more optimal management of chronic disease conditions than do existing treatments. For example, in people with coronary artery disease, visit-to-visit LDL-C variability has been shown to be an independent predictor of cardiovascular events103. Infrequent dosing intervals could be transformative for the treatment of common chronic diseases, such as hypertension and gout, for which the effectiveness of current therapies is limited by poor tolerance, compliance and prescribing practice104,105,106,107.
For indications that require deep knockdown of the target, siRNAs can be used to essentially stop expression of the bulk of the target while the remaining small fraction can be addressed with other modalities, such as antibodies. The long duration of activity, distinct mechanism of action compared with other modalities and minimal non-specific effects make RNAi therapeutics perfectly suited for combinations with other modalities. Such combinations are already being evaluated for the treatment of paroxysmal nocturnal hemoglobinuria, using a siRNA (cemdisiran) and an antibody that both target C5 (ClinicalTrials.gov: NCT04888507). This combination has the potential to minimize the amount of antibody required to inhibit C5 activity and reduce the frequency of dosing.
Multiple siRNAs can also be used in combination against different targets. With the ability of siRNAs to target extra- and intracellular proteins, including those that are difficult to drug, we anticipate that combinations of siRNAs will, for instance, be applied to simultaneously target multiple key pathways, such as those involved in tumor maintenance and growth, to reduce the likelihood of the tumor becoming resistant to therapy. They may also be useful in the treatment of cardiovascular disease, in which reduction of absolute risk drives therapeutic clinical benefit108. Following this principle, it would be preferable to simultaneously treat multiple risk factors of cardiovascular disease, such as hypertension and hypercholesterolemia. The proven safety and efficacy of the GalNAc–siRNA platform against multiple targets, and the effectiveness of its components in combination, suggests that a therapy comprising two siRNAs—for instance, one for high blood pressure and one for high cholesterol—could become a safe and effective treatment for people with metabolic syndrome. A combination approach could also be particularly beneficial against viral targets, because targeting two distinct sites will minimize the potential for resistance109.
Targets beyond the liver
We and others have recently shown robust and durable RNAi activity in several extrahepatic tissues, including the CNS, eye, muscle and tumors65,110,111. Delivering siRNA to tissues beyond the liver is the next frontier, and several such conjugates have advanced into clinical development. These include AOC-1001, a siRNA conjugated to a humanized antibody recognizing the transferrin receptor 1 for muscle targeting (ClinicalTrials.gov: NCT05027269); ALN-APP, the first CNS-targeted siRNA conjugate (ClinicalTrials.gov: NCT05231785); and ARO-HIF2, a siRNA conjugate using integrin-targeting peptides for targeted delivery to tumors. The latter advanced to mid-stage clinical development (ClinicalTrials.gov: NCT04169711), but the trial has since been terminated.
Infrequent dosing is particularly desirable for challenging routes of administration, such as intrathecal or intravitreal injections. The prolonged action of advanced RNAi therapies could therefore differentiate them from other therapeutic modalities with a shorter duration of effect. Nevertheless, non-hepatic tissues present formidable technical challenges, including efficient delivery to the target cells, efficient endosomal release and safety of the RNAi drugs and their delivery systems. The first example of targeted delivery to lung, ARO-ENaC, which combined a ligand with the epithelial integrin αvβ6 conjugated to a siRNA against the epithelial sodium channel (ENaC), was halted during early clinical development and ultimately terminated owing to toxicity observed in chronic preclinical studies. However, it should be noted that the same sponsor is now conducting clinical trials for other lung targets (ARO-MUC5AC (ClinicalTrials.gov: NCT05292950) and ARO-RAGE (ClinicalTrials.gov: NCT05276570)), with promising early results.
New target opportunities revealed by human genetics
We expect the identification of new genetically validated targets for RNAi therapeutics to accelerate, owing to the generation of ever larger genotype–phenotype datasets. The Human Genome Project (HGP) was completed at around the same time as RNAi was discovered, and both breakthroughs required many years of additional technological development to harness their full potential. The HGP’s potential was cultivated by means of affordable, high-throughput genotyping and sequencing technologies, the collection of phenotypic data from large cohorts and the ability to investigate rare genetic variants. Genome-wide association studies were intially less fruitful than anticipated because common diseases were found to be highly polygenic, with common variants having very weak effects112. Only years later did the advent of very large, genotyped case–control groups and population-scale studies enable the association of novel, more rare variants with common diseases113,114,115. Armed with insights from these results for validating therapeutic targets116,117, genetics efforts have since expanded to larger datasets, such as the UK Biobank, which collected data from 500,000 participants118,119. These and similar efforts are helping to identify suitable targets for RNAi therapeutics, as well as to predict safety issues associated with drug targets120,121. We expect this trend to continue, because larger datasets should help to uncover associations for rare loss-of-function variants and identify novel targets for RNAi and other therapeutics122.
RNAi and gene therapy
How do RNAi and ASOs compare with gene therapy and gene editing, which can offer engraftment of a curative gene or deletion of a disease-causing gene after a single dose123,124,125? It is difficult to predict their full therapeutic potential, but gene therapy and gene editing may face several limitations that prevent broad applications. First, they act more like a switch in individual cells than a rheostat, and for certain targets, near-complete suppression could have deleterious effects126—especially for non-secreted targets127,128. Second, permanent gene editing would be difficult to reverse if the need arose. In new draft guidelines, the US FDA has set a 15-year follow-up period for therapeutic gene editing and edited products (https://www.fda.gov/regulatory-information/search-fda-guidance-documents/human-gene-therapy-products-incorporating-human-genome-editing). In this regard, the RNAi approach compares favorably. The PD effects of even the longest acting siRNAs are not permanent60,129 and can potentially be reversed89. Finally, the size and metabolic instability of mRNA drugs currently limits in vivo delivery options for gene-editing technologies to complex supramolecular systems and, hence, largely to the liver and perhaps a few other highly fenestrated tissues. However, these concerns may be addressed in time.
Conclusion
After two decades of technological development, RNAi therapeutics exhibit potential for extraordinary specificity, a long duration of effect and a reversible mechanism. Their advantages include modularity and reproducibility, a growing human safety experience with multiple regulatory approvals and numerous ongoing clinical studies, and a distinct mechanism of action that is suitable for combination therapies. Translation of RNAi therapeutics from animals to humans has proved relatively predictable, leading to high success rates for clinical development. At Alnylam Pharmaceuticals, the track record is now a >60% probability of success from Investigational New Drug application to a positive phase 3 trial, compared with historical industry metrics of <14% (ref. 130).
Although today’s approved RNAi therapeutics focus on silencing disease-causing gene transcripts in the liver, efforts to expand delivery to other tissues are rapidly increasing. This will unlock many opportunities for treatment of other human diseases with a large, unmet medical need. With six approved therapies and more likely on the way, RNAi therapeutics are now firmly established as a new modality. Yet, given the potential of the technology and the vast number of targets and tissues still to be drugged, we are likely in just the earliest days of this new class of medicines.
References
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Elbashir, S. M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001).
Statello, L., Guo, C.-J., Chen, L.-L. & Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 (2021).
Ozata, D. M., Gainetdinov, I., Zoch, A., O’Carroll, D. & Zamore, P. D. PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 20, 89–108 (2019).
Baek, D. et al. The impact of microRNAs on protein output. Nature 455, 64–71 (2008).
Obbard, D. J., Gordon, K. H., Buck, A. H. & Jiggins, F. M. The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 99–115 (2009).
Crooke, S. T., Witztum, J. L., Bennett, C. F. & Baker, B. F. RNA-targeted therapeutics. Cell Metab. 27, 714–739 (2018).
Dominski, Z. & Kole, R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl Acad. Sci. USA 90, 8673–8677 (1993).
Gregory, R. I. et al. Human RISC couples MicroRNA biogenesis and posttranscriptional gene silencing. Cell 123, 631–640 (2005).
Zamore, P. D., Tuschl, T., Sharp, P. A. & Bartel, D. P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33 (2000).
Hammond, S. M., Bernstein, E., Beach, D. & Hannon, G. J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296 (2000).
Roberts, T. C. The microRNA machinery. Adv. Exp. Med. Biol. 887, 15–30 (2015).
Guo, H., Ingolia, N. T., Weissman, J. S. & Bartel, D. P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 (2010).
Eichhorn, S. W. et al. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 56, 104–115 (2014).
Liu, J. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 (2004).
Haley, B. & Zamore, P. D. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 11, 599–606 (2004).
Brown, C. R. et al. Investigating the pharmacodynamic durability of GalNAc–siRNA conjugates. Nucleic Acids Res. 48, 11827–11844 (2020).
Paunovska, K., Loughrey, D. & Dahlman, J. E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 23, 265–280 (2022).
Whitehead, K. A., Langer, R. & Anderson, D. G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 8, 129–138 (2009).
Bowie, A. G. & Unterholzner, L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 8, 911–922 (2008).
Bumcrot, D., Manoharan, M., Koteliansky, V. & Sah, D. W. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2, 711–719 (2006).
Titze-de-Almeida, R., David, C. & Titze-de-Almeida, S. S. The race of 10 synthetic RNAi-based drugs to the pharmaceutical market. Pharm. Res. 34, 1339–1363 (2017).
Garba, A. O. & Mousa, S. A. Bevasiranib for the treatment of wet, age-related macular degeneration. Ophthalmol. Eye Dis. 2, 75–83 (2010).
Eisenstein, M. Pharma’s roller-coaster relationship with RNA therapies. Nature 574, S4–S6 (2019).
Bailey, A. L. & Cullis, P. R. Modulation of membrane fusion by asymmetric transbilayer distributions of amino lipids. Biochemistry 33, 12573–12580 (1994).
Semple, S. C. et al. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: formation of novel small multilamellar vesicle structures. Biochim. Biophys. Acta 1510, 152–166 (2001).
Hafez, I. M., Maurer, N. & Cullis, P. R. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 8, 1188–1196 (2001).
Zimmermann, T. S. et al. RNAi-mediated gene silencing in non-human primates. Nature 441, 111–114 (2006).
Judge, A. D., Bola, G., Lee, A. C. & MacLachlan, I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 13, 494–505 (2006).
Semple, S. C. et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 28, 172–176 (2010).
Akinc, A. et al. Development of lipidoid–siRNA formulations for systemic delivery to the liver. Mol. Ther. 17, 872–879 (2009).
Love, K. T. et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl Acad. Sci. USA 107, 1864–1869 (2010).
Akinc, A. et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 18, 1357–1364 (2010).
Jayaraman, M. et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. Engl. 51, 8529–8533 (2012).
Cullis, P. R. & Hope, M. J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 25, 1467–1475 (2017).
Fitzgerald, K. et al. Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. Lancet 383, 60–68 (2014).
Coelho, T. et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 369, 819–829 (2013).
Adams, D. et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 11–21 (2018).
Hou, X., Zaks, T., Langer, R. & Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1074–1094 (2021).
Szabó, G. T., Mahiny, A. J. & Vlatkovic, I. COVID-19 mRNA vaccines: platforms and current developments. Mol. Ther. 30, 1850–1868 (2022).
Maier, M. A. et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 21, 1570–1578 (2013).
Sabnis, S. et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 26, 1509–1519 (2018).
Zhang, X. et al. Biodegradable amino-ester nanomaterials for Cas9 mRNA delivery in vitro and in vivo. ACS Appl. Mater. Interfaces 9, 25481–25487 (2017).
Allerson, C. R. et al. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 48, 901–904 (2005).
Morrissey, D. V. et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology 41, 1349–1356 (2005).
Nair, J. K. et al. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 136, 16958–16961 (2014).
Butler, J. S. et al. Preclinical evaluation of RNAi as a treatment for transthyretin-mediated amyloidosis. Amyloid 23, 109–118 (2016).
Stockert, R. J., Morell, A. G. & Scheinberg, I. H. Mammalian hepatic lectin. Science 186, 365–366 (1974).
Steer, C. J. & Ashwell, G. Studies on a mammalian hepatic binding protein specific for asialoglycoproteins. Evidence for receptor recycling in isolated rat hepatocytes. J. Biol. Chem. 255, 3008–3013 (1980).
Stockert, R. J. et al. Endocytosis of asialoglycoprotein-enzyme conjugates by hepatocytes. Lab Invest. 43, 556–563 (1980).
Schwartz, A. L., Fridovich, S. E. & Lodish, H. F. Kinetics of internalization and recycling of the asialoglycoprotein receptor in a hepatoma cell line. J. Biol. Chem. 257, 4230–4237 (1982).
Rensen, P. C., van Leeuwen, S. H., Sliedregt, L. A., van Berkel, T. J. & Biessen, E. A. Design and synthesis of novel N-acetylgalactosamine-terminated glycolipids for targeting of lipoproteins to the hepatic asialoglycoprotein receptor. J. Med. Chem. 47, 5798–5808 (2004).
Huang, X., Leroux, J. C. & Castagner, B. Well-defined multivalent ligands for hepatocytes targeting via asialoglycoprotein receptor. Bioconjug. Chem. 28, 283–295 (2017).
Rajeev, K. G. et al. Hepatocyte-specific delivery of siRNAs conjugated to novel non-nucleosidic trivalent N-acetylgalactosamine elicits robust gene silencing in vivo. ChemBioChem 16, 903–908 (2015).
Matsuda, S. et al. siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem. Biol. 10, 1181–1187 (2015).
Zimmermann, T. S. et al. Clinical proof of concept for a novel hepatocyte-targeting GalNAc–siRNA conjugate. Mol. Ther. 25, 71–78 (2017).
Judge, D. P. et al. Phase 3 multicenter study of revusiran in patients with hereditary transthyretin-mediated (hATTR) amyloidosis with cardiomyopathy (ENDEAVOUR). Cardiovasc. Drugs Ther. 34, 357–370 (2020).
Nair, J. K. et al. Impact of enhanced metabolic stability on pharmacokinetics and pharmacodynamics of GalNAc–siRNA conjugates. Nucleic Acids Res. 45, 10969–10977 (2017).
Foster, D. J. et al. Advanced siRNA designs further improve in vivo performance of GalNAc–siRNA conjugates. Mol. Ther. 26, 708–717 (2018).
Habtemariam, B. A. et al. Single-dose pharmacokinetics and pharmacodynamics of transthyretin targeting N-acetylgalactosamine-small interfering ribonucleic acid conjugate, vutrisiran, in healthy subjects. Clin. Pharmacol. Ther. 109, 372–382 (2021).
Ray, K. K. et al. Inclisiran in patients at high cardiovascular risk with elevated LDL cholesterol. N. Engl. J. Med. 376, 1430–1440 (2017).
Janas, M. M. et al. Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun. 9, 723 (2018).
Prakash, T. P. et al. Identification of metabolically stable 5′-phosphate analogs that support single-stranded siRNA activity. Nucleic Acids Res. 43, 2993–3011 (2015).
Parmar, R. et al. 5′-(E)-Vinylphosphonate: a stable phosphate mimic can improve the RNAi activity of siRNA-GalNAc conjugates. ChemBioChem 17, 985–989 (2016).
Brown, K. M. et al. Expanding RNAi therapeutics to extrahepatic tissues with lipophilic conjugates. Nat. Biotechnol. 40, 1500–1508 (2022).
Janas, M. M. et al. The nonclinical safety profile of GalNAc-conjugated RNAi therapeutics in subacute studies. Toxicol. Pathol. 46, 735–745 (2018).
McDougall, R. et al. The nonclinical disposition and pharmacokinetic/pharmacodynamic properties of N-acetylgalactosamine-conjugated small interfering RNA are highly predictable and build confidence in translation to human. Drug Metab. Dispos. 50, 781–797 (2022).
Larsson, E., Sander, C. & Marks, D. mRNA turnover rate limits siRNA and microRNA efficacy. Mol. Syst. Biol. 6, 433 (2010).
Agarwal, S. et al. Pharmacokinetics and pharmacodynamics of the small interfering ribonucleic acid, givosiran, in patients with acute hepatic porphyria. Clin. Pharmacol. Ther. 108, 63–72 (2020).
Hu, B. et al. Therapeutic siRNA: state of the art. Signal Transduct. Target. Ther. 5, 101 (2020).
Springer, A. D. & Dowdy, S. F. GalNAc–siRNA conjugates: leading the way for delivery of RNAi therapeutics. Nucleic Acid Ther. 28, 109–118 (2018).
Kawasaki, A. M. et al. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem. 36, 831–841 (1993).
Tobin, K. A. Macugen treatment for wet age-related macular degeneration. Insight 31, 11–14 (2006).
McKenzie, R. et al. Hepatic failure and lactic acidosis due to fialuridine (FIAU), an investigational nucleoside analogue for chronic hepatitis B. N. Engl. J. Med. 333, 1099–1105 (1995).
Richardson, F. C. et al. An evaluation of the toxicities of 2′-fluorouridine and 2′-fluorocytidine-HCl in F344 rats and woodchucks (Marmota monax). Toxicol. Pathol. 27, 607–617 (1999).
Saleh, A. F. et al. 2′-O-(2-methoxyethyl) nucleosides are not phosphorylated or incorporated into the genome of human lymphoblastoid TK6 cells. Toxicol. Sci. 163, 70–78 (2018).
Shen, W., Liang, X. H., Sun, H. & Crooke, S. T. 2′-Fluoro-modified phosphorothioate oligonucleotide can cause rapid degradation of P54nrb and PSF. Nucleic Acids Res. 43, 4569–4578 (2015).
Janas, M. M. et al. Safety evaluation of 2′-deoxy-2′-fluoro nucleotides in GalNAc–siRNA conjugates. Nucleic Acids Res. 47, 3306–3320 (2019).
Ray, K. K. et al. Long-term efficacy and safety of inclisiran in patients with high cardiovascular risk and elevated LDL cholesterol (ORION-3): results from the 4-year open-label extension of the ORION-1 trial. Lancet Diabetes Endocrinol. 11, 109–119 (2023).
Vosberg, H. P. & Eckstein, F. Effect of deoxynucleoside phosphorothioates incorporated in DNA on cleavage by restriction enzymes. J. Biol. Chem. 257, 6595–6599 (1982).
Geary, R. S., Yu, R. Z. & Levin, A. A. Pharmacokinetics of phosphorothioate antisense oligodeoxynucleotides. Curr. Opin. Investig. Drugs 2, 562–573 (2001).
Burgers, P. M. J. & Eckstein, F. Synthesis of dinucleoside monophosphorothioates via addition of sulphur to phosphite triesters. Tetrahedron Lett. 19, 3835–3838 (1978).
Crooke, S. T., Seth, P. P., Vickers, T. A. & Liang, X. H. The interaction of phosphorothioate-containing RNA targeted drugs with proteins is a critical determinant of the therapeutic effects of these Agents. J. Am. Chem. Soc. 142, 14754–14771 (2020).
Frazier, K. S. Antisense oligonucleotide therapies: the promise and the challenges from a toxicologic pathologist’s perspective. Toxicol. Pathol. 43, 78–89 (2015).
Janas, M. M. et al. Exposure to siRNA–GalNAc conjugates in systems of the standard test battery for genotoxicity. Nucleic Acid Ther. 26, 363–371 (2016).
Ray, K. K. et al. Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N. Engl. J. Med. 382, 1507–1519 (2020).
Valdmanis, P. N. et al. RNA interference-induced hepatotoxicity results from loss of the first synthesized isoform of microRNA-122 in mice. Nat. Med. 22, 557–562 (2016).
Grimm, D. et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441, 537–541 (2006).
Zlatev, I. et al. Reversal of siRNA-mediated gene silencing in vivo. Nat. Biotechnol. 36, 509–511 (2018).
Jackson, A. L. et al. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21, 635–637 (2003).
Jackson, A. L. et al. Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA 12, 1179–1187 (2006).
Birmingham, A. et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3, 199–204 (2006).
Bramsen, J. B. et al. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 38, 5761–5773 (2010).
Vaish, N. et al. Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res. 39, 1823–1832 (2011).
Schlegel, M. K. et al. From bench to bedside: improving the clinical safety of GalNAc–siRNA conjugates using seed-pairing destabilization. Nucleic Acids Res. 50, 6656–6670 (2022).
Gane, E. et al. Evaluation of RNAi therapeutics VIR-2218 and ALN-HBV for chronic hepatitis B: results from randomized clinical trials. J. Hepatol. 79, 924–932 (2023).
Zhang, M. M., Bahal, R., Rasmussen, T. P., Manautou, J. E. & Zhong, X. B. The growth of siRNA-based therapeutics: updated clinical studies. Biochem. Pharmacol. 189, 114432 (2021).
Grimm, D. Asymmetry in siRNA design. Gene Ther. 16, 827–829 (2009).
Biscans, A. et al. The chemical structure and phosphorothioate content of hydrophobically modified siRNAs impact extrahepatic distribution and efficacy. Nucleic Acids Res. 48, 7665–7680 (2020).
Kim, D. H. et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 23, 222–226 (2005).
Zhang, X., Goel, V. & Robbie, G. J. Pharmacokinetics of patisiran, the first approved RNA interference therapy in patients with hereditary transthyretin-mediated amyloidosis. J. Clin. Pharmacol. 60, 573–585 (2019).
Setten, R. L., Rossi, J. J. & Han, S. P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 18, 421–446 (2019).
Bangalore, S., Breazna, A., DeMicco, D. A., Wun, C.-C. & Messerli, F. H. Visit-to-visit low-density lipoprotein cholesterol variability and risk of cardiovascular outcomes. J. Am. Coll. Cardiol. 65, 1539–1548 (2015).
Fields, T. R. The challenges of approaching and managing gout. Rheum. Dis. Clin. North Am. 45, 145–157 (2019).
Flack, J. M. Epidemiology and unmet needs in hypertension. J. Manag. Care Pharm. 13, 2–8 (2007).
Ceral, J. et al. Difficult-to-control arterial hypertension or uncooperative patients? The assessment of serum antihypertensive drug levels to differentiate non-responsiveness from non-adherence to recommended therapy. Hypertens. Res. 34, 87–90 (2011).
Tomaszewski, M. et al. High rates of non-adherence to antihypertensive treatment revealed by high-performance liquid chromatography-tandem mass spectrometry (HP LC–MS/MS) urine analysis. Heart 100, 855–861 (2014).
Nelson, M. R. Moving the goalposts for blood pressure—time to act. N. Engl. J. Med. 385, 1328–1329 (2021).
Balakrishnan, K. N. et al. Multiple gene targeting siRNAs for down regulation of Immediate Early-2 (Ie2) and DNA polymerase genes mediated inhibition of novel rat cytomegalovirus (strain All-03). Virol. J. 17, 164 (2020).
Alterman, J. F. et al. A divalent siRNA chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous system. Nat. Biotechnol. 37, 884–894 (2019).
Chernikov, I. V., Vlassov, V. V. & Chernolovskaya, E. L. Current development of siRNA bioconjugates: from research to the clinic. Front. Pharmacol. 10, 444 (2019).
Visscher, P. M. et al. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017).
Bellenguez, C. et al. A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics 28, 134–135 (2012).
Jonsson, T. et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 488, 96–99 (2012).
Helgadottir, A. et al. Variants with large effects on blood lipids and the role of cholesterol and triglycerides in coronary disease. Nat. Genet. 48, 634–639 (2016).
Plenge, R. M., Scolnick, E. M. & Altshuler, D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 12, 581–594 (2013).
King, E. A., Davis, J. W. & Degner, J. F. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 15, e1008489 (2019).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Szustakowski, J. D. et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat. Genet. 53, 942–948 (2021).
Nguyen, P. A., Born, D. A., Deaton, A. M., Nioi, P. & Ward, L. D. Phenotypes associated with genes encoding drug targets are predictive of clinical trial side effects. Nat. Commun. 10, 1579 (2019).
Diogo, D. et al. Phenome-wide association studies across large population cohorts support drug target validation. Nat. Commun. 9, 4285 (2018).
Minikel, E. V. et al. Evaluating drug targets through human loss-of-function genetic variation. Nature 581, 459–464 (2020).
Khan, F. A. et al. CRISPR/Cas9 therapeutics: a cure for cancer and other genetic diseases. Oncotarget 7, 52541–52552 (2016).
Herrera-Carrillo, E., Gao, Z. & Berkhout, B. CRISPR therapy towards an HIV cure. Brief. Funct. Genomics 19, 201–208 (2020).
Gillmore, J. D. et al. CRISPR–Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502 (2021).
Wittrup, A. & Lieberman, J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 16, 543–552 (2015).
Egeberg, O. Thrombophilia caused by inheritable deficiency of blood antithrombin. Scand. J. Clin. Lab. Invest. 17, 92 (1965).
Pasi, K. J. et al. Targeting of antithrombin in hemophilia A or B with RNAi therapy. N. Engl. J. Med. 377, 819–828 (2017).
Fitzgerald, K. et al. A highly durable RNAi therapeutic inhibitor of PCSK9. N. Engl. J. Med. 376, 41–51 (2017).
Wong, C. H., Siah, K. W. & Lo, A. W. Estimation of clinical trial success rates and related parameters. Biostatistics 20, 273–286 (2019).
Chan, A. et al. Preclinical development of a subcutaneous ALAS1 RNAi therapeutic for treatment of hepatic porphyrias using circulating RNA quantification. Mol. Ther. Nucleic Acids 4, e263 (2015).
Balwani, M. et al. Phase 3 trial of RNAi therapeutic givosiran for acute intermittent porphyria. N. Engl. J. Med. 382, 2289–2301 (2020).
Garrelfs, S. F. et al. Lumasiran, an RNAi therapeutic for primary hyperoxaluria type 1. N. Engl. J. Med. 384, 1216–1226 (2021).
Wright, R. S. et al. Pooled patient-level analysis of inclisiran trials in patients with familial hypercholesterolemia or atherosclerosis. J. Am. Coll. Cardiol. 77, 1182–1193 (2021).
Adams, D. et al. Efficacy and safety of vutrisiran for patients with hereditary transthyretin-mediated amyloidosis with polyneuropathy: a randomized clinical trial. Amyloid 30, 1–9 (2023).
Zhu, Y. et al. RNA-based therapeutics: an overview and prospectus. Cell Death Dis. 13, 644 (2022).
Singh, R. N. & Singh, N. N. Mechanism of splicing regulation of spinal muscular atrophy genes. Adv. Neurobiol. 20, 31–61 (2018).
Ackermann, E. J. et al. Suppressing transthyretin production in mice, monkeys and humans using 2nd-generation antisense oligonucleotides. Amyloid 23, 148–157 (2016).
Bartel, D. P. Metazoan microRNAs. Cell 173, 20–51 (2018).
Napoli, C., Lemieux, C. & Jorgensen, R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 2, 279–289 (1990).
Soutschek, J. et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 432, 173–178 (2004).
Acknowledgements
We thank the many contributors at Alnylam and other colleagues in the field who have contributed to the advancement of oligonucleotide therapeutics over the past several decades. We offer special thanks to T. Auperin for his tremendous support during the writing and editing of this manuscript. We also acknowledge the medical writing services provided by E. Childs and colleagues of Adelphi Communications (Macclesfield, UK), in accordance with Good Publication Practice guidelines, funded by Alnylam Pharmaceuticals.
Author information
Authors and Affiliations
Contributions
V.J., A.V., K.F. and M.A.M. outlined and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
All authors are or have been employees of Alnylam Pharmaceuticals during the writing of the manuscript and report ownership of equity in Alnylam Pharmaceuticals: V.J. is chief technology officer; A.V. is chief innovation officer; K.F. is chief scientific officer; and M.A.M. is senior vice-president, head of oncology.
Peer review
Peer review information
Nature Biotechnology thanks Arthur Krieg and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jadhav, V., Vaishnaw, A., Fitzgerald, K. et al. RNA interference in the era of nucleic acid therapeutics. Nat Biotechnol 42, 394–405 (2024). https://doi.org/10.1038/s41587-023-02105-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-023-02105-y