Abstract 抽象
There has been considerable recent progress in designing new proteins using deep-learning methods1,2,3,4,5,6,7,8,9. Despite this progress, a general deep-learning framework for protein design that enables solution of a wide range of design challenges, including de novo binder design and design of higher-order symmetric architectures, has yet to be described. Diffusion models10,11 have had considerable success in image and language generative modelling but limited success when applied to protein modelling, probably due to the complexity of protein backbone geometry and sequence–structure relationships. Here we show that by fine-tuning the RoseTTAFold structure prediction network on protein structure denoising tasks, we obtain a generative model of protein backbones that achieves outstanding performance on unconditional and topology-constrained protein monomer design, protein binder design, symmetric oligomer design, enzyme active site scaffolding and symmetric motif scaffolding for therapeutic and metal-binding protein design. We demonstrate the power and generality of the method, called RoseTTAFold diffusion (RFdiffusion), by experimentally characterizing the structures and functions of hundreds of designed symmetric assemblies, metal-binding proteins and protein binders. The accuracy of RFdiffusion is confirmed by the cryogenic electron microscopy structure of a designed binder in complex with influenza haemagglutinin that is nearly identical to the design model. In a manner analogous to networks that produce images from user-specified inputs, RFdiffusion enables the design of diverse functional proteins from simple molecular specifications.
最近在使用深度学习方法 1,2,3,4,5,6,7,8,9 设计新蛋白质方面取得了相当大的进展。尽管取得了这些进展,但尚未描述用于蛋白质设计的通用深度学习框架,该框架能够解决广泛的设计挑战,包括从头结合剂设计和高阶对称结构的设计。扩散模型 10,11 在图像和语言生成建模方面取得了相当大的成功,但在应用于蛋白质建模时成功有限,这可能是由于蛋白质骨架几何形状和序列-结构关系的复杂性。在这里,我们展示了通过微调RoseTTAFold结构预测网络在蛋白质结构去噪任务上,我们得到了一个蛋白质骨架的生成模型,该模型在无条件和拓扑约束的蛋白质单体设计、蛋白质结合剂设计、对称寡聚物设计、酶活性位点支架和对称基序支架上取得了优异的性能,用于治疗和金属结合蛋白质设计。我们通过实验表征数百种设计的对称组装体、金属结合蛋白和蛋白质结合剂的结构和功能,证明了该方法的强大性和通用性,称为 RoseTTAFold 扩散 (RFdiffusion)。RF扩散的准确性通过设计粘合剂与流感血凝素复合物的低温电子显微镜结构得到证实,该粘合剂与设计模型几乎相同。RFdiffusion类似于从用户指定的输入生成图像的网络,能够从简单的分子规格设计出不同的功能蛋白。
Similar content being viewed by others
其他人正在查看的类似内容
Main 主要
De novo protein design seeks to generate proteins with specified structural and/or functional properties, for example, making a binding interaction with a given target12, folding into a particular topology13 or containing a catalytic site4. Denoising diffusion probabilistic models (DDPMs), a powerful class of machine learning models recently demonstrated to generate new photorealistic images in response to text prompts14,15, have several properties well suited to protein design. First, DDPMs generate highly diverse outputs, as they are trained to denoise data (for instance, images or text) that have been corrupted with Gaussian noise. By learning to stochastically reverse this corruption, diverse outputs closely resembling the training data are generated. Second, DDPMs can be guided at each step of the iterative generation process towards specific design objectives through provision of conditioning information. Third, for almost all protein design applications it is necessary to explicitly model three-dimensional (3D) structures; rotationally equivariant DDPMs can do this in a global representation frame independent manner. Recent work has adapted DDPMs for protein monomer design by conditioning on small protein ‘motifs’5,9 or on secondary structure and block-adjacency (‘fold’) information8. Although promising, these attempts have shown limited success in generating sequences that fold to the intended structures in silico5,16, probably due to the limited ability of the denoising networks to generate realistic protein backbones, and have not been tested experimentally.
从头蛋白质设计旨在生成具有特定结构和/或功能特性的蛋白质,例如,与给定靶标 12 进行结合相互作用,折叠成特定的拓扑结构 13 或包含催化位点 4 。去噪扩散概率模型 (DDPM) 是一类强大的机器学习模型,最近被证明可以生成新的逼真图像 14,15 以响应文本提示,具有多种非常适合蛋白质设计的特性。首先,DDPM生成高度多样化的输出,因为它们经过训练,可以对被高斯噪声破坏的数据(例如,图像或文本)进行降噪。通过学习随机逆转这种腐败,可以生成与训练数据非常相似的各种输出。其次,通过提供调节信息,可以在迭代生成过程的每个步骤中引导DDPM实现特定的设计目标。第三,对于几乎所有的蛋白质设计应用,都需要显式地对三维(3D)结构进行建模;旋转等变 DDPM 可以以独立于全局表示帧的方式执行此操作。最近的工作通过调节小蛋白质“基序” 5,9 或二级结构和块邻接(“折叠”)信息 8 来调整DDPMs进行蛋白质单体设计。尽管很有希望,但这些尝试在生成在计算机中折叠到预期结构的序列方面显示出有限的成功 5,16 ,这可能是由于去噪网络产生真实蛋白质骨架的能力有限,并且尚未进行实验测试。
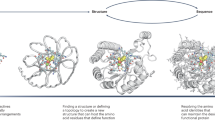
We reasoned that improved diffusion models for protein design could be developed by taking advantage of the deep understanding of protein structure implicit in powerful structure prediction methods such as AlphaFold2 (ref. 17) (AF2) and RoseTTAFold18 (RF). RF has properties well suited for use in a protein design DDPM (Fig. 1a): it generates protein structures with high precision, operates on a rigid-frame representation of residues with rotational equivariance and has an architecture enabling conditioning on design specifications at the individual residue, inter-residue distance and orientation, and 3D coordinate levels. In previous work, we fine-tuned RF to complete protein backbones around input functional motifs in a single step (RFjoint Inpainting4). Experimental characterization showed that the method can scaffold a wide range of protein functional motifs with atomic accuracy19, but the approach fails on minimalist site descriptions that do not sufficiently constrain the overall fold and, because it is deterministic, can produce only a limited diversity of designs for a given problem. We reasoned that by fine-tuning RF as the denoising network in a generative diffusion model instead, we could overcome both problems: because the starting point is random noise, each denoising trajectory yields a different solution, and because structure is built up progressively through many denoising iterations, little to no starting structural information should be required. In this study, we used an updated version of RF18 as the basis for the denoising network architecture (Supplementary Methods), but other equivariant structure prediction networks (AF2 (ref. 17), OmegaFold20, ESMFold21) could in principle be substituted into an analogous DDPM.
我们推断,通过利用强大的结构预测方法(如AlphaFold2(参考文献 17 )(AF2)和RoseTTAFold 18 (RF))中隐含的对蛋白质结构的深刻理解,可以开发用于蛋白质设计的改进扩散模型。RF 具有非常适合用于蛋白质设计 DDPM(图 1a)的特性:它以高精度生成蛋白质结构,在具有旋转等方差的残基的刚性框架表示上运行,并且具有能够在单个残基、残基间距离和方向以及 3D 坐标水平上调节设计规范的架构。在之前的工作中,我们对 RF 进行了微调,以在单个步骤中完成围绕输入功能基序的蛋白质骨架 (RF joint Inpainting 4 )。实验表征表明,该方法可以以原子精度搭建广泛的蛋白质功能基序 19 ,但该方法在极简主义位点描述上失败了,这些位点描述不能充分约束整体折叠,并且由于它是确定性的,只能为给定的问题产生有限的设计多样性。我们推断,通过在生成扩散模型中微调RF作为去噪网络,我们可以克服这两个问题:因为起点是随机噪声,所以每个去噪轨迹都会产生不同的解决方案,并且由于结构是通过许多去噪迭代逐步建立起来的,因此几乎不需要起始结构信息。在这项研究中,我们使用更新版本的RF 18 作为去噪网络架构(补充方法)的基础,但其他等变结构预测网络(AF2(参考文献)。 17 )、OmegaFold 20 、ESMFold 21 )原则上可以替换为类似的DDPM。
a, Diffusion models for proteins are trained to recover corrupted (noised) protein structures and to generate new structures by reversing the corruption process through iterative denoising of initially random noise XT into a realistic structure X0 (top panel). The RF structure prediction network (middle panel, left side) is fine-tuned with minimal architectural changes into RFdiffusion (middle panel, right side); the denoising network of a DDPM is also shown. In RF, the primary input to the model is the sequence. In RFdiffusion, the primary input is diffused residue frames (coordinates and orientations). In both cases, the model predicts final 3D coordinates (denoted in RFdiffusion). The bottom panel shows that in RFdiffusion, the model receives its previous prediction as a template input (‘self-conditioning’, Supplementary Methods). At each timestep t of a trajectory (typically 200 steps), RFdiffusion takes from the previous step and Xt and then predicts an updated X0 structure (). The next coordinate input to the model () is generated by a noisy interpolation (interp) towards . b, RFdiffusion is broadly applicable for protein design. RFdiffusion generates protein structures either without further input (top row) or by conditioning on (top to bottom): symmetry specifications; binding targets; protein functional motifs or symmetric functional motifs. In each case random noise, along with conditioning information, is input to RFdiffusion, which iteratively refines that noise until a final protein structure is designed. c, An example of an unconditional design trajectory for a 300-residue chain, depicting the input to the model (Xt) and the corresponding prediction. At early timesteps (high t), bears little resemblance to a protein but is gradually refined into a realistic protein structure.
a,蛋白质的扩散模型经过训练,可以恢复损坏的(噪声)蛋白质结构,并通过将初始随机噪声 X T 迭代去噪到真实结构 X 0 中来逆转损坏过程来生成新结构(上图)。RFdiffusion(中间面板,右侧)对RFdiffusion(中间面板,右侧)进行了微调,架构变化最小;还显示了DDPM的去噪网络。在 RF 中,模型的主要输入是序列。在 RFdiffusion 中,主要输入是扩散残差帧(坐标和方向)。在这两种情况下,模型都会预测最终的 3D 坐标(用 RFdiffusion 表示 )。底部面板显示,在 RFdiffusion 中,模型接收其先前的预测作为模板输入(“自调节”,补充方法)。在轨迹的每个时间步长 t(通常为 200 步)处,RFdiffusion 从前一步和 X t 获取 ,然后预测更新的 X 0 结构 ( )。模型 ( ) 的下一个坐标输入由朝向 的噪声插值 (interp) 生成。b,RF扩散广泛适用于蛋白质设计。RFdiffusion 无需进一步输入(顶行)或通过调节(从上到下)生成蛋白质结构:对称规范;约束目标;蛋白质功能基序或对称功能基序。在每种情况下,随机噪声以及条件信息都会被输入到RFdiffusion中,RFdiffusion会迭代细化该噪声,直到设计出最终的蛋白质结构。 c,300 个残基链的无条件设计轨迹示例,描述了模型 (X t ) 的输入和相应的 预测。在早期时间步长(高t), 与蛋白质几乎没有相似之处,但逐渐被细化为现实的蛋白质结构。
We construct a RF-based diffusion model, RFdiffusion, using the RF frame representation that comprises a Cα coordinate and N-Cα-C rigid orientation for each residue. We generate training inputs by noising structures sampled from the Protein Data Bank (PDB) for up to 200 steps22. For translations, we perturb Cα coordinates with 3D Gaussian noise. For residue orientations, we use Brownian motion on the manifold of rotation matrices (building on refs. 23,24). To enable RFdiffusion to learn to reverse each step of the noising process, we train the model by minimizing a mean-squared error (m.s.e.) loss between frame predictions and the true protein structure (without alignment), averaged across all residues (Supplementary Methods). This loss drives denoising trajectories to match the data distribution at each timestep and hence to converge on structures of designable protein backbones (Extended Data Fig. 2a). The m.s.e. contrasts to the loss used in RF structure prediction training (frame aligned point error or FAPE) in that, unlike FAPE, m.s.e. loss is not invariant to the global reference frame and therefore promotes continuity of the global coordinate frame between timesteps (Supplementary Methods).
我们构建了一个基于RF的扩散模型RFdiffusion,使用RF框架表示,该帧表示包括每个残基的Cα坐标和N-Cα-C刚性取向。我们通过对从蛋白质数据库 (PDB) 采样的结构进行多达 200 步 22 的噪声来生成训练输入。对于平移,我们用 3D 高斯噪声扰动 Cα 坐标。对于残基取向,我们在旋转矩阵的流形上使用布朗运动(建立在 refs. 23,24 )。为了使RFdiffusion能够学习逆转噪声过程的每一步,我们通过最小化帧预测和真实蛋白质结构(无比对)之间的均方误差(m.s.e.)损失来训练模型,平均所有残基(补充方法)。这种损失驱动去噪轨迹以匹配每个时间步长的数据分布,从而收敛于可设计蛋白质骨架的结构(扩展数据图2a)。m.s.e. 与射频结构预测训练中使用的损耗(帧对准点误差或 FAPE)形成鲜明对比,因为与 FAPE 不同,m.s.e. 损耗不是全局参考系的不变的,因此促进了时间步长之间全局坐标系的连续性(补充方法)。
To generate a new protein backbone, we first initialize random residue frames and RFdiffusion makes a denoised prediction. Each residue frame is updated by taking a step in the direction of this prediction with some noise added to generate the input to the next step. The nature of the noise added and the size of this reverse step is chosen such that the denoising process matches the distribution of the noising process (Supplementary Methods and Extended Data Fig. 2a). RFdiffusion initially seeks to match the full breadth of possible protein structures compatible with the purely random frames with which it is initialized, and hence the denoised structures do not initially seem protein-like (Fig. 1c, left). However, through many such steps, the breadth of possible protein structures from which the input could have arisen narrows and RFdiffusion predictions come to closely resemble protein structures (Fig. 1c, right). We use the ProteinMPNN network1 to subsequently design sequences encoding these structures, typically sampling eight sequences per design in line with previous work5,16 (but see Supplementary Fig. 2a). We also considered simultaneously designing structure and sequence within RFdiffusion, but given the excellent performance of combining ProteinMPNN with the diffusion of structure alone, we did not extensively explore this possibility.
为了生成新的蛋白质骨架,我们首先初始化随机残基框架,然后RFdiffusion进行去噪预测。每个残差帧都通过朝着这个预测的方向迈出一步来更新,并添加一些噪声以生成下一步的输入。选择添加的噪声的性质和该反向步骤的大小,使得去噪过程与噪声过程的分布相匹配(补充方法和扩展数据图2a)。RFdiffusion 最初寻求匹配与其初始化的纯随机框架兼容的可能蛋白质结构的全部宽度,因此去噪结构最初看起来不像蛋白质(图 1c,左)。然而,通过许多这样的步骤,可能产生输入的蛋白质结构的广度变窄了,RF扩散预测变得与蛋白质结构非常相似(图1c,右)。我们使用 ProteinMPNN 网络 1 随后设计编码这些结构的序列,通常每个设计采样 8 个序列,这与以前的工作 5,16 一致(但参见补充图 2a)。我们还考虑在RFdiffusion中同时设计结构和序列,但鉴于将ProteinMPNN与单独结构扩散相结合的优异性能,我们没有广泛探索这种可能性。
Figure 1a highlights the similarities between RF structure prediction and an RFdiffusion denoising step: in both cases, the networks transform coordinates into a predicted structure, conditioned on inputs to the model. In RF, sequence is the primary input, with extra structural information provided as templates and initial coordinates to the model. In RFdiffusion, the primary input is the noised coordinates from the previous step. For specific design tasks, a range of auxiliary conditioning information, including partial sequence, fold information or fixed functional-motif coordinates can be provided (Fig. 1b and Supplementary Methods).
图 1a 突出显示了 RF 结构预测和 RF扩散去噪步骤之间的相似之处:在这两种情况下,网络都将坐标转换为预测结构,条件是模型的输入。在 RF 中,序列是主要输入,额外的结构信息作为模板和初始坐标提供给模型。在 RFdiffusion 中,主要输入是上一步中的噪声坐标。对于特定的设计任务,可以提供一系列辅助调节信息,包括部分序列、折叠信息或固定的功能基序坐标(图 1b 和补充方法)。
We explored two different strategies for training RFdiffusion: (1) in a manner akin to ‘canonical’ diffusion models, with predictions at each timestep independent of predictions at previous timesteps (as in previous work5,8,9,16), and (2) with self-conditioning25, in which the model can condition on previous predictions between timesteps (Fig. 1a, bottom row and Supplementary Methods). The latter strategy was inspired by the success of ‘recycling’ in AF2, which is also central to the more recent RF model used here (Supplementary Methods). Self-conditioning within RFdiffusion notably improved performance on in silico benchmarks encompassing both conditional and unconditional protein design tasks (Fig. 2e and Extended Data Fig. 1e). Increased coherence of predictions within self-conditioned trajectories may, at least in part, explain these performance increases (Extended Data Fig. 1h). Fine-tuning RFdiffusion from pretrained RF weights was far more successful than training for an equivalent length of time from untrained weights (Extended Data Fig. 1f,g, also Supplementary Fig. 1) and the m.s.e. loss was also crucial for unconditional generation (Extended Data Fig. 1d). For all in silico benchmarks in this paper, we use the AF2 structure prediction network17 for validation and define an in silico ‘success’ as an RFdiffusion output for which the AF2 structure predicted from a single sequence is (1) of high confidence (mean predicted aligned error (pAE), less than five), (2) globally within a 2 Å backbone root mean-squared deviation (r.m.s.d.) of the designed structure and (3) within 1 Å backbone r.m.s.d. on any scaffolded functional site (Supplementary Methods). This measure of in silico success has been found to correlate with experimental success4,7,26 and is significantly more stringent than template modelling (TM)-score-based metrics used elsewhere5,16,27,28,29 (Supplementary Fig. 2c,d).
我们探索了两种不同的训练RFdiffusion的策略:(1)以类似于“规范”扩散模型的方式,每个时间步的预测独立于先前时间步的预测(如以前的工作 5,8,9,16 ),以及(2)使用自调节 25 ,其中模型可以根据时间步之间的先前预测进行条件(图1a,底行和补充方法)。后一种策略的灵感来自AF2中“回收”的成功,这也是这里使用的最新射频模型(补充方法)的核心。RFdiffusion中的自调节显著提高了计算机基准测试的性能,包括条件和无条件蛋白质设计任务(图2e和扩展数据图1e)。在自我调节轨迹中,预测的连贯性增加可能至少部分解释了这些性能的提高(扩展数据图1h)。从预训练的RF权重中微调RF扩散比从未经训练的权重中训练相同时间长度的RF扩散要成功得多(扩展数据图1f,g,也是补充图1),并且m.s.e.损失对于无条件生成也至关重要(扩展数据图1d)。对于本文中的所有计算机基准,我们使用 AF2 结构预测网络 17 进行验证,并将计算机“成功”定义为 RF扩散输出,其中从单个序列预测的 AF2 结构为 (1) 高置信度(平均预测对齐误差 (pAE),小于 5),(2) 在设计结构的 2 Å 主干根均方差 (r.m.s.d.) 内,以及 (3) 在任何支架上 1 Å 主干 r.m.s.d. 内功能站点(补充方法)。 已发现这种计算机成功衡量标准与实验成功 4,7,26 相关,并且比其他地方使用的基于模板建模 (TM) 分数的指标要严格得多 5,16,27,28,29 (补充图 2c、d)。
Unconditional protein monomer generation
无条件蛋白质单体生成
As shown in Fig. 2a–c and Supplementary Fig. 3c,d, starting from random noise, RFdiffusion can readily generate elaborate protein structures with little overall structural similarity to structures seen during training, indicating considerable generalization beyond the PDB (see Supplementary Table 1 for a comparison of all designs in the paper to the PDB). The designs are diverse (Supplementary Fig. 3a), spanning a wide range of alpha, beta and mixed alpha–beta topologies, with AF2 and ESMFold (Fig. 2c, Extended Data Fig. 1b,c and Supplementary Fig. 2b) predictions very close to the design structure models for de novo designs with as many as 600 residues. RFdiffusion generates plausible structures for even very large proteins, but these are difficult to validate in silico as they are probably generally beyond the single sequence prediction capabilities of AF2 and ESMFold. The quality and diversity of designs that are sampled are inherent to the model, and do not depend on any auxiliary conditioning input (for example, secondary structure information8). We experimentally characterized six of the 300 amino acid designs and three of the 200 amino acid designs, and found that they have circular dichroism spectra consistent with the mixed alpha–beta topologies of the designs and are extremely thermostable (Extended Data Fig. 3). Physics-based protein design methodologies have struggled in unconstrained generation of diverse protein monomers because of the difficulty of sampling on the very large and rugged conformational landscape30, and overcoming this limitation has been a primary test of deep-learning based protein design approaches5,6,8,16,27,31. RFdiffusion strongly outperforms (based on the AF2 success metric described above) Hallucination with RF, an experimentally validated method using Monte Carlo search or gradient descent to identify sequences predicted to fold into stable structures (Fig. 2d). RFdiffusion generation is also more compute efficient than unconstrained Hallucination with RF, and efficiency can be greatly improved by taking larger steps at inference time and by truncating trajectories early, which is possible because RF predicts the final structure at each timestep (Extended Data Fig. 2b,c). For example, a 100-residue protein can be generated in as little as 11 s on an NVIDIA RTX A4000 Graphical Processing Unit, in contrast to RF Hallucination, which takes around 8.5 min.
如图2a-c和补充图3c,d所示,从随机噪声开始,RFdiffusion可以很容易地产生复杂的蛋白质结构,与训练期间看到的结构几乎没有整体结构相似性,这表明PDB之外有相当大的泛化(参见补充表1,将论文中的所有设计与PDB进行比较)。这些设计是多种多样的(补充图3a),涵盖了广泛的α、β和混合α-β拓扑结构,AF2和ESMFold(图2c,扩展数据图1b,c和补充图2b)的预测非常接近具有多达600个残基的从头设计的设计结构模型。RFdiffusion甚至可以为非常大的蛋白质生成合理的结构,但这些结构很难在计算机中验证,因为它们通常可能超出了AF2和ESMFold的单序列预测能力。抽样设计的质量和多样性是模型固有的,不依赖于任何辅助条件输入(例如,二级结构信息 8 )。我们对 300 个氨基酸设计中的 6 个和 200 个氨基酸设计中的 3 个进行了实验表征,发现它们具有与设计的混合 α-β 拓扑一致的圆二色谱,并且具有极高的热稳定性(扩展数据图 3)。基于物理的蛋白质设计方法在不受限制地生成不同的蛋白质单体方面一直很困难,因为在非常大和崎岖的构象景观 30 上进行采样的困难,克服这一限制一直是基于深度学习的蛋白质设计方法 5,6,8,16,27,31 的主要测试。 RFdiffusion 的性能优于(基于上述 AF2 成功指标)RF 幻觉,这是一种经过实验验证的方法,使用蒙特卡洛搜索或梯度下降来识别预测折叠成稳定结构的序列(图 2d)。RF扩散生成也比使用RF的无约束幻觉具有更高的计算效率,并且通过在推理时采取更大的步骤和尽早截断轨迹可以大大提高效率,这是可能的,因为RF在每个时间步预测最终结构(扩展数据图2b,c)。例如,在 NVIDIA RTX A4000 图形处理单元上,可以在短短 11 秒内生成 100 个残基的蛋白质,而射频幻觉则需要大约 8.5 分钟。
图2:RF扩散在单体生成中的出色性能。
a, RFdiffusion can generate new monomeric proteins of different lengths (left 300, right 600) with no conditioning information. Grey, design model; colours, AF2 prediction. r.m.s.d. AF2 versus design (Å), left to right: 0.90, 0.98, 1.15, 1.67. b, Unconditional designs from RFdiffusion are new and not present in the training set as quantified by highest TM-score to the PDB; the divergence from previously known structures increases with length. c, Unconditional samples are closely repredicted by AF2 up to about 400 amino acids. d, RFdiffusion significantly outperforms Hallucination (with RF) at unconditional monomer generation (two-proportion z-test of in silico success: n = 400 designs per condition, z = 9.5, P = 1.6 × 10−21). Although Hallucination successfully generates designs up to 100 amino acids in length, in silico success rates rapidly deteriorate beyond this length. e, Ablating pretraining (by starting from untrained RF), RFdiffusion fine-tuning (that is, using original RF structure prediction weights as the denoiser), self-conditioning or m.s.e. losses (by training with FAPE) each notably decrease the performance of RFdiffusion. r.m.s.d. between design and AF2 is shown, for the unconditional generation of 300 amino acid proteins (Supplementary Methods). f, Two example 300 amino acid proteins that expressed as soluble monomers. Designs (grey) overlaid with AF2 predictions (colours) are shown on the left, alongside circular dichroism (CD) spectra (top) and melt curves (bottom) on the right. The designs are highly thermostable. g, RFdiffusion can condition on fold information. An example TIM barrel is shown (bottom left), conditioned on the secondary structure and block adjacency of a previously designed TIM barrel, PDB 6WVS (top left). Designs have very similar circular dichroism spectra to PDB 6WVS (top right) and are highly thermostable (bottom right). See also Extended Data Fig. 3 for further traces. Boxplots represent median ± interquartile range; tails are minimum and maximum excluding outliers (±1.5× interquartile range).
a,RFdiffusion可以产生不同长度(左300,右600)的新单体蛋白,无需条件信息。灰色,设计模型;颜色,AF2 预测。r.m.s.d. AF2 与设计 (Å),从左到右:0.90、0.98、1.15、1.67。b,RFdiffusion 的无条件设计是新的,并且不存在于训练集中,由 PDB 的最高 TM 分数量化;与先前已知结构的差异随着长度的增加而增加。c,无条件样品通过AF2密切重新预测,最多可达约400个氨基酸。d,RF扩散在无条件单体生成时明显优于幻觉(使用RF)(计算机成功的双比例z检验:每个条件n = 400个设计,z = 9.5,P = 1.6 × 10 −21 )。尽管幻觉成功地产生了长达100个氨基酸的设计,但在计算机模拟中,超过这个长度的成功率会迅速下降。e、消融预训练(从未训练的射频开始)、射频扩散微调(即使用原始射频结构预测权重作为降噪器)、自调节或 m.s.e. 损失(通过使用 FAPE 训练)都会显著降低射频扩散的性能。r.m.s.d. 在设计和 AF2 之间,用于无条件生成 300 个氨基酸蛋白质(补充方法)。f,两种以可溶性单体形式表达的300个氨基酸的实例。左侧显示叠加了 AF2 预测(颜色)的设计(灰色),右侧显示了圆二色性 (CD) 光谱(上)和熔体曲线(下)。这些设计具有高度的热稳定性。g, RF扩散可以调节折叠信息。图中显示了一个示例 TIM 枪管(左下角),该枪管以先前设计的 TIM 枪管 PDB 6WVS(左上角)的二级结构和块相邻性为条件。 该设计具有与PDB 6WVS(右上)非常相似的圆二色光谱,并且具有高度的热稳定性(右下)。有关进一步的迹线,另请参阅扩展数据图 3。箱线图表示四分位距±位距的中位数;尾部是最小值和最大值,不包括异常值(±1.5×四分位距)。
It is often desirable to be able to specify a protein fold during design (such as triose-phosphate isomerase (TIM) barrels or cavity-containing NTF2s for small molecule binder and enzyme design32,33), and thus we further fine-tuned RFdiffusion to condition on secondary structure and/or fold information, enabling rapid and accurate generation of diverse designs with the desired topologies (Fig. 2g and Extended Data Fig. 4). In silico success rates were 42.5 and 54.1% for TIM barrels and NTF2 folds, respectively (Extended Data Fig. 4d), and experimental characterization of 11 TIM barrel designs indicated that at least eight designs were soluble, thermostable and had circular dichroism spectra consistent with the design model (Fig. 2g and Extended Data Fig. 4e,f).
通常希望能够在设计过程中指定蛋白质折叠(例如用于小分子结合剂和酶设计的 32,33 磷酸丙糖异构酶 (TIM) 桶或含腔的 NTF2),因此我们进一步微调 RFdiffusion 以调节二级结构和/或折叠信息,从而能够快速准确地生成具有所需拓扑结构的不同设计(图 2g 和扩展数据图 4)。TIM桶和NTF2折叠的计算机成功率分别为42.5%和54.1%(扩展数据图4d),对11种TIM桶设计的实验表征表明,至少有8种设计是可溶的、热稳定的,并且具有与设计模型一致的圆二色光谱(图2g和扩展数据图4e,f)。
Design of higher-order oligomers
高阶低聚物的设计
There is considerable interest in designing symmetric oligomers, which can serve as vaccine platforms34, delivery vehicles35 and catalysts36. Cyclic oligomers have been designed using structure prediction networks with an adaptation of Hallucination that searches for sequences predicted to fold to the desired cyclic symmetry, but this approach fails for higher-order dihedral, tetrahedral, octahedral and icosahedral symmetries, probably in part because of the much lower representation of such structures in the PDB7.
人们对设计对称低聚物有相当大的兴趣,这些低聚物可以用作疫苗平台 34 、运载工具 35 和催化剂 36 。环状低聚物是使用结构预测网络设计的,该网络具有幻觉的适应性,该网络搜索预测为折叠到所需循环对称性的序列,但这种方法对于高阶二面体、四面体、八面体和二十面体对称性无效,部分原因可能是此类结构在 PDB 7 中的表示性要低得多。
We set out to generalize RFdiffusion to create symmetric oligomeric structures with any specified point group symmetry. Given a specification of a point group symmetry for an oligomer with n chains, and the monomer chain length, we generate random starting residue frames for a single monomer subunit as in the unconditional generation case, and then generate n − 1 copies of this starting point arranged with the specified point group symmetry. Because RFdiffusion is equivariant (inherited from RF) with respect to rotation and relabelings of chains, symmetry is largely maintained in the denoising predictions; we explicitly resymmetrize at each step but this changes the structures only slightly (compare grey and coloured chains in Extended Data Fig. 5a and Supplementary Methods). For octahedral and icosahedral architectures, we explicitly model only the smallest subset of monomers required to generate the full assembly (for example, for icosahedra, the subunits at the five-, three- and twofold symmetry axes) to reduce the computational cost and memory footprint.
我们开始推广RFdiffusion,以创建具有任何指定点群对称性的对称低聚结构。给定具有 n 条链的低聚物的点群对称性规范和单体链长度,我们为单个单体亚基生成随机起始残基框架,如无条件生成情况,然后生成该起点的 n − 1 个副本以指定的点群对称性排列。由于RF扩散在链的旋转和重新标记方面是等变的(继承自RF),因此在去噪预测中在很大程度上保持了对称性;我们在每个步骤中都明确地进行了再对称,但这仅略微改变了结构(比较扩展数据图5a和补充方法中的灰色和彩色链)。对于八面体和二十面体架构,我们仅明确模拟生成完整组装所需的最小单体子集(例如,对于二十面体,五重、三重和二重对称轴处的亚基),以降低计算成本和内存占用。
Despite not being trained on symmetric inputs, RFdiffusion is able to generate symmetric oligomers with high in silico success rates (Extended Data Fig. 5b), particularly when guided by an auxiliary inter- and intrachain contact potential (Extended Data Fig. 5c). As illustrated in Fig. 3 and Extended Data Fig. 5e, RFdiffusion designs are nearly indistinguishable from AF2 predictions of the structures adopted by the designed sequences, and many show little resemblance to previously solved protein structures (Extended Data Fig. 5d and Supplementary Table 1). Several of the oligomeric topologies are not seen in the PDB, including two-layer beta barrels (Fig. 3a, C10 symmetry) and complex mixed alpha/beta topologies (Fig. 3a, C8 symmetry; closest TM align in PDB 6BRP, 0.47, and PDB 6BRO, 0.43, respectively).
尽管没有经过对称输入的训练,但RFdiffusion能够产生具有高计算机成功率的对称低聚物(扩展数据图5b),特别是在辅助链间和链内接触电位的引导下(扩展数据图5c)。如图3和扩展数据图5e所示,RF扩散设计与所设计序列所采用的结构的AF2预测几乎没有区别,并且许多与先前解析的蛋白质结构几乎没有相似之处(扩展数据图5d和补充表1)。PDB中没有几种低聚拓扑结构,包括两层β桶(图3a,C10对称性)和复杂的混合α/β拓扑结构(图3a,C8对称性;最接近的TM对齐分别在PDB 6BRP(0.47)和PDB 6BRO(0.43)中)。
图3:对称低聚物的设计和实验表征。
a, RFdiffusion-generated assemblies overlaid with the AF2 structure predictions based on the designed sequences; in all five cases they are nearly indistinguishable (for the octahedron (bottom), the prediction was for the C3 substructure). Symmetries are indicated to the left of the design models. b,c, Designed assemblies characterized by nsEM. Model symmetries are as follows: cyclic, C3 (HE0822, 350 amino acids (AA) per chain), C6 (HE0626, 100 AA per chain) and C8 (HE0675, 60 AA per chain) (b); dihedral, D3 (HE0490, 80 AA per chain) and D4 (HE0537, 100 AA per chain) (c). From left to right: (1) symmetric design model, (2) AF2 prediction of design following sequence design with ProteinMPNN, (3) 2D class averages showing both top and side views (scale bar, 60 Å for all class averages) and (4) 3D reconstructions from class averages with the design model fit into the density map. The overall shapes are consistent with the design models, and confirm the intended oligomeric state. As in a, AF2 predictions of each design are nearly indistinguishable from the design model (backbone r.m.s.d.s (Å) for HE0822, HE0626, HE0490, HE0675 and HE0537, are 1.33, 1.03, 0.60, 0.74 and 0.75, respectively). d, nsEM characterization of an icosahedral particle (HE0902, 100 AA per chain). The design model, including the AF2 prediction of the C3 subunit are shown on the left. nsEM data are shown on the right: on top, a representative micrograph is shown alongside 2D class averages along each symmetry axis (C3, C2 and C5, from left to right) with the corresponding 3D reconstruction map views shown directly below overlaid on the design model.
a,RFdiffusion生成的组件叠加基于设计序列的AF2结构预测;在所有五种情况下,它们几乎无法区分(对于八面体(底部),预测是针对C3子结构的)。对称性指示在设计模型的左侧。b,c, 以 nsEM 为特征的设计组件。模型对称性如下:环状,C3(HE0822,每链350个氨基酸(AA)),C6(HE0626,每链100个AA)和C8(HE0675,每链60个AA)(b);二面体,D3(HE0490,每链 80 AA)和 D4(HE0537,每链 100 AA)(c)。从左到右:(1)对称设计模型,(2)使用ProteinMPNN进行序列设计后设计的AF2预测,(3)显示顶视图和侧视图的2D类平均值(比例尺,所有类平均值为60 Å)和(4)使用设计模型拟合密度图的类平均值进行3D重建。整体形状与设计模型一致,并确认了预期的低聚状态。与a一样,每种设计的AF2预测与设计模型几乎没有区别(HE0822、HE0626、HE0490、HE0675和HE0537的骨干r.m.s.d.s(Å)分别为1.33、1.03、0.60、0.74和0.75)。d,二十面体颗粒的nsEM表征(HE0902,每链100 AA)。设计模型,包括 C3 亚基的 AF2 预测如左图所示。nsEM 数据显示在右侧:在顶部,代表性的显微照片与沿每个对称轴(C3、C2 和 C5,从左到右)的 2D 类平均值一起显示,相应的 3D 重建图视图直接显示在设计模型上。
We selected 608 designs for experimental characterization and found using size-exclusion chromatography (SEC) that at least 87 had oligomerization states closely consistent with the design models (within the 95% confidence interval, 126 designs within the 99% confidence interval, as determined by SEC calibration curves; Supplementary Figs. 4 and 5). We took advantage of the increased size of these oligomers (compared to the smaller unconditional and fold-conditioned monomers described above) and collected negative stain electron microscopy (nsEM) data on a subset of these designs across different symmetry groups. For most, distinct particles were evident with shapes resembling the design models in both the raw micrographs and subsequent two-dimensional (2D) classifications (Fig. 3 and Extended Data Fig. 5f). nsEM characterization of a C3 design (HE0822) with 350 residue subunits (1,050 residues in total) suggests that the actual structure is very close to the design, both over the 350 residue subunits and the overall C3 architecture. 2D class averages are clearly consistent with both top and side views of the design model, and a 3D reconstruction of the density has key features consistent with the design, including the distinctive pinwheel shape (Fig. 3b, top row). Electron microscopy 2D class averages of C5 and C6 designs with more than 750 residues (HE0794, HE0789, HE0841) were also consistent with the respective design models (Extended Data Fig. 5f).
我们选择了 608 个设计进行实验表征,并使用尺寸排阻色谱 (SEC) 发现至少有 87 个设计具有与设计模型密切相关的寡聚化状态(在 95% 置信区间内,126 个设计在 99% 置信区间内,由 SEC 校准曲线确定;补充图4和图5)。我们利用了这些低聚物的尺寸增加(与上述较小的无条件和折叠条件单体相比),并收集了不同对称组中这些设计子集的负染色电子显微镜 (nsEM) 数据。对于大多数人来说,在原始显微照片和随后的二维(2D)分类中,不同的颗粒具有明显的形状,其形状与设计模型相似(图3和扩展数据图5f)。具有 350 个残基亚基(总共 1,050 个残基)的 C3 设计 (HE0822) 的 nsEM 表征表明,实际结构与设计非常接近,无论是超过 350 个残基亚基还是整个 C3 结构。2D 等级平均值与设计模型的顶视图和侧视图明显一致,密度的 3D 重建具有与设计一致的关键特征,包括独特的风车形状(图 3b,顶行)。具有超过 750 个残基的 C5 和 C6 设计(HE0794、HE0789、HE0841)的电子显微镜 2D 类平均值也与各自的设计模型一致(扩展数据图 5f)。
RFdiffusion also generated cyclic oligomers with alpha and/or beta barrel structures that resemble expanded TIM barrels and provide an interesting comparison between innovation during natural evolution and innovation through deep learning. The TIM barrel fold, with eight strands and eight helices, is one of the most abundant folds in nature37. nsEM confirmed the structure of two RFdiffusion designed cyclic oligomers, which considerably extend beyond this fold (Fig. 3b, bottom rows). HE0626 is a C6 alpha–beta barrel composed of 18 strands and 18 helices, and HE0675 is a C8 octamer composed of an inner ring of 16 strands and an outer ring of 16 helices arranged locally in a very similar repeating pattern to the TIM barrel (1:1 helix:strand). For both HE0626 and HE0675 we obtained nsEM 3D reconstructions that are in agreement with the computational design models. The HE0600 design is also an alpha–beta barrel (Extended Data Fig. 5f), but has two strands for every helix (24 strands and 12 helices in total) and hence is locally different from a TIM barrel. Whereas natural evolution has extensively explored structural variations of the classic eight-strand or eight-helix TIM barrel fold, RFdiffusion can more readily explore global changes in barrel curvature, enabling discovery of TIM barrel-like structures with many more helices and strands.
RFdiffusion 还生成了具有 α 和/或 β 桶结构的环状低聚物,类似于膨胀的 TIM 桶,并在自然进化过程中的创新与通过深度学习的创新之间进行了有趣的比较。TIM桶形褶皱有八股和八个螺旋,是自然界 37 中最丰富的褶皱之一。nsEM证实了两种RFdiffusion设计的环状低聚物的结构,它们大大超出了这个折叠(图3b,底部行)。HE0626 是由 18 条链和 18 个螺旋组成的 C6 α-β 桶,HE0675 是一种 C8 八聚体,由 16 条链的内环和 16 个螺旋的外环组成,以与 TIM 桶非常相似的重复模式局部排列(1:1 螺旋:股)。对于 HE0626 和 HE0675,我们获得了与计算设计模型一致的 nsEM 3D 重建。HE0600 设计也是一个 α-β 桶(扩展数据图 5f),但每个螺旋有两条链(总共 24 条链和 12 个螺旋),因此在局部上与 TIM 桶不同。虽然自然进化已经广泛探索了经典的八链或八螺旋TIM桶状褶皱的结构变化,但RFdiffusion可以更容易地探索桶曲率的全局变化,从而能够发现具有更多螺旋和链的TIM桶状结构。
RFdiffusion also readily generated structures with dihedral, tetrahedral and icosohedral symmetries (Fig. 3c,d and Extended Data Fig. 5e,f). SEC characterization indicated that 38 D2, seven D3 and three D4 designs had the expected molecular weights (these have four, six and eight chains, respectively) (Supplementary Fig. 5). Although the D2 dihedrals are too small for nsEM, 2D class averages—and for some, 3D reconstructions of D3 and D4 designs—were congruent with the overall topologies of the design models (Fig. 3c and Extended Data Fig. 5f). Similarly, 3D reconstruction (Fig. 3c) and cryogenic electron microscopy (cryo-EM) 2D class averages (Extended Data Fig. 5g and Supplementary Fig. 6) of the D4 HE0537 closely match the design model, recapitulating the roughly 45° offset between tetramic subunits. 2D nsEM class averages for a 12-chain tetrahedron (HE0964) were consistent with the design model (Extended Data Fig. 5f). Forty-eight icosahedra were selected for experimental validation, and one, HE0902, a 15 nm (diameter) highly porous assembly (Fig. 3d, left) was observed in nsEM micrographs to form homogeneous particles. 2D class averages and a 3D reconstruction very closely match the design model (Fig. 3d), with triangular hubs arrayed around the empty C5 axes. Designs such as HE0902 (and future similar large assemblies) should be useful as new nanomaterials and vaccine scaffolds, with robust assembly and (in the case of HE0902) the outward facing N and C termini offering many possibilities for antigen display.
RF扩散也很容易产生具有二面体、四面体和二十面体对称性的结构(图3c,d和扩展数据图5e,f)。SEC表征表明,38个D2、7个D3和3个D4设计具有预期的分子量(它们分别有4条、6条和8条链)(补充图5)。尽管 D2 二面体对于 nsEM 来说太小,但 2D 类平均值(对于某些人来说,D3 和 D4 设计的 3D 重建)与设计模型的整体拓扑结构一致(图 3c 和扩展数据图 5f)。同样,D4 HE0537 的 3D 重建(图 3c)和低温电子显微镜 (cryo-EM) 2D 类平均值(扩展数据图 5g 和补充图 6)与设计模型非常吻合,概括了四亚基之间大约 45° 的偏移。12链四面体(HE0964)的二维nsEM类平均值与设计模型一致(扩展数据图5f)。选择了 48 个二十面体进行实验验证,其中一个是 HE0902,在 nsEM 显微照片中观察到一个 15 nm(直径)高多孔组件(Fig. 3d,左)以形成均匀颗粒。2D 类平均值和 3D 重建与设计模型 (Fig. 3d) 非常匹配,三角形轮毂围绕空的 C5 轴排列。HE0902(以及未来类似的大型组件)等设计应该可以用作新的纳米材料和疫苗支架,具有坚固的组件和(在HE0902的情况下)朝外的N和C末端,为抗原显示提供了许多可能性。
Functional-motif scaffolding
功能主题脚手架
We next investigated the use of RFdiffusion for scaffolding protein structural motifs that carry out binding and catalytic functions, in which the role of the scaffold is to hold the motif in precisely the 3D geometry needed for optimal function. In RFdiffusion, we input motifs as 3D coordinates (including sequence and sidechains) both during conditional training and inference, and build scaffolds that hold the motif atomic coordinates in place. Many deep-learning methods have been developed recently to address this problem, including RFjoint Inpainting4, constrained Hallucination4 and other DDPMs5,8,29. To rigorously evaluate the performance of these methods in comparison to RFdiffusion across a broad set of design challenges, we established an in silico benchmark test (Supplementary Table 9) comprising 25 motif-scaffolding design problems addressed in six recent publications encompassing several design methodologies4,5,29,38,39,40. The challenges span a broad range of motifs, including simple ‘inpainting’ problems, viral epitopes, receptor traps, small molecule binding sites, binding interfaces and enzyme active sites.
接下来,我们研究了RFdiffusion在执行结合和催化功能的蛋白质结构基序支架中的应用,其中支架的作用是将基序精确地保持在最佳功能所需的3D几何形状中。在RFdiffusion中,我们在条件训练和推理过程中将基序作为3D坐标(包括序列和侧链)输入,并构建将基序原子坐标固定到位的支架。最近已经开发了许多深度学习方法来解决这个问题,包括 RF joint Inpainting 4 、约束幻觉 4 和其他 DDPM。 5,8,29 为了严格评估这些方法与RFdiffusion相比,在一系列广泛的设计挑战中的性能,我们建立了一个计算机基准测试(补充表9),其中包括25个主题脚手架设计问题,这些问题在最近的六份出版物中得到解决,包括多种设计方法 4,5,29,38,39,40 。这些挑战涵盖了广泛的基序,包括简单的“修复”问题、病毒表位、受体陷阱、小分子结合位点、结合界面和酶活性位点。
RFdiffusion solves 23 of the 25 benchmark problems, compared to 15 for Hallucination and 19 for RFjoint Inpainting (Fig. 4a,b). For 19 out of 23 of the problems solved by RFdiffusion, the fraction of successful designs is higher than either Hallucination or RFjoint Inpainting. The excellent performance of RFdiffusion required no hyperparameter tuning or external potentials; this contrasts with Hallucination, for which problem-specific optimization can be required. In 17 out of 23 of the problems, RFdiffusion-generated successful solutions with higher in silico success rates when noise was not added during the reverse diffusion trajectories (see Extended Data Fig. 1i for further discussion on the effect of noise on design quality, and Supplementary Fig. 8 for analysis of design diversity). The ability of RFdiffusion to scaffold functional motifs is not related to their presence in the RFdiffusion training set (Supplementary Fig. 7).
RFdiffusion 解决了 25 个基准问题中的 23 个,而幻觉为 15 个,RF joint 修复为 19 个(图 4a,b)。在RFdiffusion解决的23个问题中,有19个问题的成功设计比例高于幻觉或RF joint 修复。RFdiffusion 的出色性能不需要超参数调整或外部电位;这与幻觉形成鲜明对比,幻觉可能需要针对特定问题的优化。在23个问题中的17个问题中,当在反向扩散轨迹中未添加噪声时,RFdiffusion生成的成功解具有更高的计算机成功率(有关噪声对设计质量影响的进一步讨论,请参阅扩展数据图1i,以及有关设计多样性分析的补充图8)。RFdiffusion 支架功能基序的能力与它们在 RFdiffusion 训练集中的存在无关(补充图 7)。
图4:使用RFdiffusion构建不同功能位点的支架。
a, RFdiffusion outperforms other methods across 25 benchmark motif-scaffolding problems collected from six recent publications (Supplementary Table 9). In silico success is defined as AF2 r.m.s.d. to design model less than 2 Å, AF2 r.m.s.d. to the native functional motif less than 1 Å and AF2 pAE less than five. One hundred designs were generated per problem, with no previous optimization on the benchmark set (some optimization was necessary for Hallucination). Supplementary Table 10 presents full results. In silico success rates on the problems are correlated between the methods, and RFdiffusion can still struggle on challenging problems in which all methods have low success. b, Four examples of designs in which RFdiffusion significantly outperforms existing methods. Teal, native motif; colours, AF2 prediction of a design. Metrics (r.m.s.d. AF2 versus design/versus native motif (Å), AF2 pAE): 5TRV long, 1.17/0.57; 4.73; 6E6R long, 0.89/0.27, 4.56; 7MRX long, 0.84/0.82 4.32; 5TPN, 0.59/0.49 3.77. c, RFdiffusion can scaffold the p53 helix that binds MDM2 (left) and makes extra contacts with the target (right, average 31% increased surface area. Design was p53_design_89). Designs were generated with an RFdiffusion model fine-tuned on complexes. d, BLI measurements indicate high-affinity binding to MDM2 (p53_design_89, 0.7 nM; p53_design_53, 0.5 nM); the native affinity is 600 nM (ref. 42). e, Out of 95 designs, 55 showed binding to MDM2 (more than 50% of maximum response). Thirty-two of these were monomeric (Supplementary Fig. 10h). f, After fine-tuning (Supplementary Methods), RFdiffusion can scaffold enzyme active sites. An oxidoreductase example (EC1) is shown (PDB 1A4I); catalytic site (teal); RFdiffusion output (grey, model; colours, AF2 prediction); zoom of active site. AF2 versus design backbone r.m.s.d. 0.88 Å, AF2 versus design motif backbone r.m.s.d. 0.53 Å, AF2 versus design motif full-atom r.m.s.d. 1.05 Å, AF2 pAE 4.47. g, In silico success rates on active sites derived from EC1-5 (AF2 Motif r.m.s.d. versus native: backbone less than 1 Å, backbone and sidechain atoms less than 1.5 Å, r.m.s.d. AF2 versus design less than 2 Å, AF2 pAE less than 5).
a,RFdiffusion 在从最近的六篇出版物中收集的 25 个基准基序脚手架问题中优于其他方法(补充表 9)。计算机模拟成功定义为 AF2 r.m.s.d. 设计模型小于 2 Å,AF2 r.m.s.d. 设计小于 1 Å 的天然功能基序,AF2 pAE 小于 5。每个问题生成了 100 个设计,之前没有对基准集进行优化(幻觉需要一些优化)。补充表10列出了完整的结果。在计算机模拟中,方法之间的问题成功率是相关的,RFdiffusion在所有方法的成功率都很低的挑战性问题上仍然很困难。b,RFdiffusion明显优于现有方法的四个设计示例。蓝绿色,原生图案;颜色,设计的 AF2 预测。指标(r.m.s.d. AF2 与设计/与天然基序 (Å),AF2 pAE):5TRV 长,1.17/0.57;4.73;6E6R长,0.89/0.27,4.56;7MRX长,0.84/0.82 4.32;5TPN,0.59/0.49 3.77。c,RFdiffusion 可以支撑结合 MDM2(左)并与靶标进行额外接触的 p53 螺旋(右,表面积平均增加 31%。设计是p53_design_89)。设计是使用在复合物上微调的 RFdiffusion 模型生成的。d, BLI测量表明与MDM2的高亲和力结合(p53_design_89,0.7 nM;p53_design_53,0.5 nM);天然亲和力为 600 nM(参考文献)。 42 e,在 95 种设计中,55 种显示出与 MDM2 的结合(超过最大响应的 50%)。其中32个是单体的(补充图10h)。f、微调(补充方法)后,RF扩散可以支架酶活性位点。 示为氧化还原酶示例(EC1)(PDB 1A4I);催化位点(蓝绿色);RF扩散输出(灰色,模型;颜色,AF2预测);活动站点的缩放。AF2 与设计主链 r.m.s.d. 0.88 Å,AF2 与设计主链 r.m.s.d. 0.53 Å,AF2 与设计基序全原子 r.m.s.d. 1.05 Å,AF2 pAE 4.47。g,源自 EC1-5 的活性位点的计算机模拟成功率(AF2 Motif r.m.s.d. vs 天然:骨架小于 1 Å,主链和侧链原子小于 1.5 Å,r.m.s.d. AF2 与设计小于 2 Å,AF2 pAE 小于 5)。
One of the benchmark problems is the scaffolding of the p53 helix that binds MDM2. Inhibiting this interaction through high-affinity competitive inhibition by scaffolding the p53 helix and making further interactions with MDM2 is a promising therapeutic avenue41. In silico success has been described elsewhere4, but experimental success has not been reported. We used an RFdiffusion model fine-tuned on protein complexes (Supplementary Methods) to generate 96 designs scaffolding this helix. We scaffolded the p53 helix in the presence of MDM2, so extra interactions could be designed by RFdiffusion and experimentally identified 0.5 and 0.7 nM binders (Fig. 4c,d), three orders of magnitude higher affinity than the reported 600 nM affinity of the p53 peptide alone42. The overall success rate was quite high: out of the 96 designs, 55 showed some detectable binding at 10 μM (Fig. 4e and Supplementary Fig. 10h).
基准问题之一是结合MDM2的p53螺旋的支架。通过支架 p53 螺旋并通过高亲和力竞争性抑制来抑制这种相互作用并与 MDM2 进行进一步的相互作用是一种很有前途的治疗途径 41 。计算机模拟成功在其他地方已有描述 4 ,但实验成功尚未报道。我们使用对蛋白质复合物进行微调的 RFdiffusion 模型(补充方法)来生成 96 个设计支架。我们在 MDM2 存在下搭建了 p53 螺旋支架,因此可以通过 RFdiffusion 设计额外的相互作用,并通过实验鉴定出 0.5 和 0.7 nM 结合剂(图 4c,d),亲和力比单独 42 报道的 p53 肽的 600 nM 亲和力高三个数量级。总体成功率相当高:在96种设计中,55种在10μM处显示出一些可检测到的结合(图4e和补充图10h)。
Scaffolding enzyme active sites
支架酶活性位点
A grand challenge in protein design is to scaffold minimal descriptions of enzyme active sites comprising a few single amino acids. Whereas some in silico success has been reported previously4, a general solution that can readily produce high-quality, orthogonally validated outputs remains elusive. Following fine-tuning on a task mimicking this problem (Supplementary Methods), RFdiffusion was able to scaffold enzyme active sites comprising many sidechain and backbone functional groups with high accuracy and in silico success rates across a range of enzyme classes (Fig. 4f and Extended Data Fig. 6a–d; in silico success required fine tuning). Although RFdiffusion is unable to explicitly model bound small molecules at present (however, see our conclusions), the substrate can be implicitly modelled using an external potential to guide the generation of ‘pockets’ around the active site. As a demonstration, we scaffold a retroaldolase active site triad while implicitly modelling the reaction substrate (Extended Data Fig. 6e–h).
蛋白质设计的一大挑战是支撑由几个单个氨基酸组成的酶活性位点的最小描述。虽然之前 4 已经报道过一些计算机模拟成功,但可以很容易地产生高质量、正交验证输出的通用解决方案仍然难以捉摸。在对模拟此问题的任务进行微调(补充方法)后,RFdiffusion能够高精度地搭建包含许多侧链和骨架官能团的酶活性位点,并在一系列酶类别中实现计算机成功率(图4f和扩展数据图6a-d;计算机成功需要微调)。尽管RFdiffusion目前无法显式地模拟结合的小分子(但是,请参阅我们的结论),但可以使用外部电位隐式模拟底物,以引导活性位点周围“口袋”的产生。作为演示,我们在隐式模拟反应底物的同时搭建了反醛缩酶活性位点三联体(扩展数据图6e-h)。
Symmetric functional-motif scaffolding
对称功能主题脚手架
Several important design challenges involve the scaffolding of several copies of a functional motif in symmetric arrangements. For example, many viral glycoproteins are trimeric and symmetry matched arrangements of inhibitory domains can be extremely potent43,44,45,46. Conversely, symmetric presentation of viral epitopes in an arrangement that mimics the virus could induce new classes of neutralizing antibodies47,48. To explore this general direction, we sought to design trimeric multivalent binders to the SARS-CoV-2 spike protein. In previous work, flexible linkage of a binder to the ACE2 binding site (on the spike protein receptor binding domain) to a trimerization domain yielded a high-affinity inhibitor that had potent and broadly neutralizing antiviral activity in animal models43. Ideally, however, symmetric fusions to binders would be rigid, so as to reduce the entropic cost of binding while maintaining the avidity benefits from multivalency. We used RFdiffusion to design C3-symmetric trimers that rigidly hold three binding domains (the functional motif in this case) such that they exactly match the ACE2 binding sites on the SARS-CoV-2 spike protein trimer. The designs were confidently predicted by AF2 to both assemble as C3-symmetric oligomers, and to scaffold the AHB2 SARS-CoV-2 binder interface with high accuracy (Fig. 5a).
几个重要的设计挑战涉及以对称排列的方式搭建功能图案的多个副本的脚手架。例如,许多病毒糖蛋白是三聚体的,并且抑制结构域的对称匹配排列可能非常有效 43,44,45,46 。相反,病毒表位以模拟病毒的排列对称呈现可以诱导新类别的中和抗体 47,48 。为了探索这一大方向,我们试图设计SARS-CoV-2刺突蛋白的三聚体多价结合剂。在以前的工作中,结合剂与 ACE2 结合位点(在刺突蛋白受体结合域上)与三聚体化结构域的灵活连接产生了一种高亲和力抑制剂,该抑制剂在动物模型 43 中具有有效且广泛的中和抗病毒活性。然而,理想情况下,与粘合剂的对称融合将是刚性的,以降低结合的熵成本,同时保持多价的亲和力益处。我们使用 RFdiffusion 设计了 C3 对称三聚体,该三聚体刚性地保持三个结合域(本例中的功能基序),使它们与 SARS-CoV-2 刺突蛋白三聚体上的 ACE2 结合位点完全匹配。AF2 自信地预测这些设计既组装为 C3 对称低聚物,又可以高精度地搭建 AHB2 SARS-CoV-2 粘合剂界面(图 5a)。
图5:使用RFdiffusion的对称基序脚手架。
a, Design of symmetric oligomers scaffolding the binding interface of ACE2 mimic AHB2 (left, teal) against the SARS-CoV-2 spike trimer (left, grey). Three AHB2 copies are input to RFdiffusion along with C3 noise (middle); output are C3-symmetric oligomers holding the three AHB2 copies in place to engage all spike subunits. AF2 predictions (right) recapitulate the AHB2 structure with 0.6 Å r.m.s.d. over the assymetric unit and 2.9 Å r.m.s.d. over the C3 assembly. b, Design of C4-symmetric oligomers to scaffold a Ni2+ binding motif (left). Starting from square-planar histidine rotamers within helical fragments (Supplementary Methods), RFdiffusion generates a C4 oligomer scaffolding the binding domain (middle). AF2 predictions (colour) agree closely with the design model (grey), with backbone r.m.s.d. less than 1.0 Å (right). c, nsEM 2D class averages (scale bar, 60 Å) and 3D reconstruction density are consistent with the symmetry and structure of the NiB1.17 design model shown superimposed on the density in ribbon representation (top). Isothermal titration calorimetry binding isotherm of design NiB1.17 (blue) indicates a dissociation constant less than 20 nM at a metal:monomer stoichiometry of 1:4. The H52A mutant isotherm (pink) ablates binding, indicating scaffolded histidine residues are critical for metal binding. d, Additional experimentally characterized Ni2+ binders NiB2.15 (left), NiB1.12 (middle) and NiB1.20 (right). Metal-coordinating sidechains in the design models (top, teal) are closely recapitulated in the AF2 predictions (colours). 2D nsEM class averages (middle; scale bar, 60 Å) are consistent with design models. Binding isotherms for wild-type (WT) and H52A mutant (bottom) indicate Ni2+ binding mediated directly by the scaffolded histidines at the designed stoichiometry. Note that for ITC plots, points represent single measurements.
a,对称低聚物支架的设计,该支架将 ACE2 模拟 AHB2(左,蓝绿色)与 SARS-CoV-2 刺突三聚体(左,灰色)的结合界面。三个 AHB2 拷贝与 C3 噪声(中间)一起输入到 RFdiffusion;输出是 C3 对称低聚物,将三个 AHB2 拷贝固定到位以接合所有刺突亚基。AF2 预测(右)概括了 AHB2 结构,在非对称单元上 0.6 Å r.m.s.d.,在 C3 组件上 2.9 Å r.m.s.d.。b,C4对称低聚物的设计,以支撑Ni 2+ 结合基序(左)。从螺旋片段内的方形平面组氨酸旋转体(补充方法)开始,RFdiffusion 产生 C4 低聚物支架,该支架位于结合域(中间)。AF2 预测(彩色)与设计模型(灰色)非常吻合,主干 r.m.s.d. 小于 1.0 Å(右)。c,nsEM 2D 类平均值(比例尺,60 Å)和 3D 重建密度与 NiB1.17 设计模型的对称性和结构一致,该模型叠加在色带表示中的密度上(顶部)。设计NiB1.17(蓝色)的等温滴定量热法结合等温线表示在金属:单体化学计量比为1:4时的解离常数小于20 nM。H52A突变体等温线(粉红色)消融结合,表明支架组氨酸残基对金属结合至关重要。d, 其他实验表征的Ni 2+ 结合剂NiB2.15(左)、NiB1.12(中)和NiB1.20(右)。设计模型中的金属配位侧链(顶部、蓝绿色)在 AF2 预测(颜色)中得到了密切概括。二维 nsEM 类平均值(中间;比例尺,60 Å)与设计模型一致。 野生型 (WT) 和 H52A 突变体(底部)的结合等温线表明,在设计的化学计量下,支架组氨酸直接介导 Ni 2+ 结合。请注意,对于 ITC 图,点表示单个测量值。
The ability to scaffold functional sites with any desired symmetry opens up new approaches to designing metal-coordinating protein assemblies49,50. Divalent transition metal ions show distinct preferences for specific coordination geometries (for example, square planar, tetrahedral and octahedral) with ion-specific optimal sidechain–metal bond lengths. RFdiffusion provides a general route to building up symmetric protein assemblies around such sites, with the symmetry of the assembly matching the symmetry of the coordination geometry. As a first test, we sought to design square-planar Ni2+ binding sites. We designed C4 protein assemblies with four central histidine imidazoles arranged in an ideal Ni2+-binding site with square-planar coordination geometry (Fig. 5b). Diverse designs starting from distinct C4-symmetric histidine square-planar sites had good in silico success with the histidine residues in near ideal geometries for coordinating metal in the AF2-predicted structures (Supplementary Fig. 9).
以任何所需的对称性搭建功能位点的能力为设计金属配位蛋白组装开辟了新的方法 49,50 。二价过渡金属离子对具有离子特异性最佳侧链-金属键长度的特定配位几何形状(例如,方形平面、四面体和八面体)表现出不同的偏好。RFdiffusion 提供了一种围绕这些位点构建对称蛋白质组装体的一般途径,组装体的对称性与配位几何体的对称性相匹配。作为第一个测试,我们试图设计方形平面的Ni 2+ 结合位点。我们设计了 C4 蛋白组装体,其中四个中心组氨酸咪唑排列在理想的 Ni 2+ 结合位点,具有方形平面配位几何形状(图 5b)。从不同的 C4 对称组氨酸方形平面位点开始的多样化设计在计算机模拟中取得了良好的成功,组氨酸残基具有接近理想几何形状的配位金属 AF2 预测结构(补充图 9)。
We expressed and purified 44 designs in Escherichia coli, and found that 37 had SEC chromatograms consistent with the intended oligomeric state (Extended Data Fig. 7b). Of the designs, 36 were tested for Ni2+ coordination by isothermal titration calorimetry, and 18 were found to bind Ni2+ with dissociation constants ranging from low nanomolar to low micromolar (Fig. 5c,d and Extended Data Fig. 7a). The inflection points in the wild-type isotherms indicate binding with the designed stoichiometry, a one to four ratio of ion to monomer. Although most of the designed proteins showed exothermic metal coordination, in a few cases binding was endothermic (Fig. 5d, left and Extended Data Fig. 7a: NiB2.9, NiB2.10, NiB2.15 and NiB2.23), suggesting that Ni2+ coordination is entropically driven in these assemblies. To confirm that Ni2+ binding was indeed mediated by the scaffolded histidine 52, we mutated this residue to alanine, which abolished or notably reduced binding in 17 out of 17 cases with successful expression (Extended Data Figs. 7a,c and Fig. 5c,d; one mutant did not express). We structurally characterized by nsEM a subset of the designs—NiB1.12, NiB1.15, NiB1.17 and NiB1.20—that showed histidine-dependent binding. All four designs showed clear fourfold symmetry both in the raw micrographs and in 2D class averages (Fig. 5c,d), with design NiB1.17 also clearly showing twofold axis side views with a measured diameter approximating the design model. A 3D reconstruction of NiB1.17 was in close agreement with the design model (Fig. 5c).
我们在大肠杆菌中表达并纯化了 44 种设计,发现 37 种具有与预期低聚体状态一致的 SEC 色谱图(扩展数据图 7b)。在这些设计中,36 种通过等温滴定量热法测试了 Ni 2+ 配位,发现 18 种与 Ni 2+ 结合,解离常数范围从低纳摩尔到低微摩尔(图 5c,d 和扩展数据图 7a)。野生型等温线中的拐点表示与设计的化学计量结合,离子与单体的比例为一:四。尽管大多数设计的蛋白质显示出放热金属配位,但在少数情况下结合是吸热的(图5d,左图和扩展数据图7a:NiB2.9,NiB2.10,NiB2.15和NiB2.23),表明 2+ Ni配位在这些组装体中是熵驱动的。为了确认 Ni 2+ 结合确实是由支架组氨酸 52 介导的,我们将该残基突变为丙氨酸,在 17 例成功表达的病例中,有 17 例消除或显着减少结合(扩展数据图 7a,c 和图 5c,d;一个突变体不表达)。我们通过nsEM在结构上表征了设计的一个子集——NiB1.12、NiB1.15、NiB1.17和NiB1.20——显示出组氨酸依赖性结合。所有四种设计在原始显微照片和二维类平均值中都显示出清晰的四重对称性(图 5c,d),设计 NiB1.17 也清楚地显示了测量直径近似于设计模型的双轴侧视图。NiB1.17的3D重建与设计模型非常吻合(图5c)。
Design of protein-binding proteins
蛋白质结合蛋白的设计
The design of high-affinity binders to target proteins is a grand challenge in protein design, with numerous therapeutic applications51. A general method for de novo binder design from target structure information alone using the physically based Rosetta method was recently described12, and subsequently, using ProteinMPNN for sequence design and AF2 for design filtering was found to improve design success rates26. However, experimental success rates were low, still requiring many thousands of designs to be screened for each design campaign12, and the approach relied on prespecifying a particular set of protein scaffolds as the basis for the designs, inherently limiting the diversity and shape complementarity of possible solutions12. To our knowledge, no deep-learning method has yet demonstrated experimental general success in designing completely de novo binders.
靶蛋白的高亲和力结合剂的设计是蛋白质设计中的一大挑战,具有许多治疗应用 51 。最近描述了 12 一种使用基于物理的 Rosetta 方法仅从目标结构信息从头设计粘合剂的通用方法,随后发现使用 ProteinMPNN 进行序列设计,使用 AF2 进行设计过滤可以提高设计成功率 26 。然而,实验成功率很低,仍然需要为每个设计活动 12 筛选数千个设计,并且该方法依赖于预先指定一组特定的蛋白质支架作为设计的基础,这固有地限制了可能解决方案 12 的多样性和形状互补性。据我们所知,目前还没有一种深度学习方法在设计完全从头粘合剂方面显示出实验性的成功。
We reasoned that RFdiffusion might be able to address this challenge by directly generating binding proteins in the context of the target. For many therapeutic applications, for example, blocking a protein–protein interaction, it is desirable to bind to a particular site on a target protein. To enable this, we fine-tuned RFdiffusion on protein complex structures, providing a feature as input indicating a subset of the residues on the target chain (called ‘interface hotspots’) to which the diffused chain binds (Fig. 6a and Extended Data Fig. 8a,b). For design challenges in which a particular binder fold might be especially compatible, we enabled coarse-grained control over binder scaffold topology by fine-tuning an extra model to condition binder diffusion on secondary structure and block-adjacency information, in addition to conditioning on interface hotspots (Extended Data Fig. 8c,d and Supplementary Methods).
我们推断RFdiffusion可能能够通过在靶标的背景下直接产生结合蛋白来应对这一挑战。对于许多治疗应用,例如阻断蛋白质-蛋白质相互作用,希望与靶蛋白上的特定位点结合。为了实现这一点,我们对蛋白质复合物结构的RFdiffusion进行了微调,提供了一个特征作为输入,指示扩散链结合的靶链上残基的子集(称为“界面热点”)(图6a和扩展数据图8a,b)。对于特定粘结剂折叠可能特别兼容的设计挑战,我们通过微调额外的模型来调节粘结剂支架拓扑结构的粗粒度控制,以根据二级结构和块邻接信息调节粘结剂扩散,此外还对界面热点进行调节(扩展数据图 8c,d 和补充方法)。
图6:蛋白质结合蛋白的从头设计。
a, RFdiffusion generates protein binders given a target and specification of interface hotspot residues. b, De novo binders were designed to five protein targets; Influenza A H1 HA, IL-7Rα, InsR, PD-L1 and TrkA and hits with BLI response greater than or equal to 50% of the positive control were identified for all targets. For IL-7Rα, InsR, PD-L1 and TrkA, RFdiffusion has success rates roughly two orders of magnitude higher than the original design campaigns. We attribute one order of magnitude to RFdiffusion, and the second to filtering with AF2 (estimated success rates for previous campaigns if AF2 filtering had been used: HA, 0%; IL-7Rα, 2.2%; InsR, 5.5%; PD-L1, 3.7%; TrkA, 1.5%). c, For IL-7Rα, InsR, PD-L1 and TrkA, the highest affinity binder is shown above a BLI titration series. Reported KD values are based on global kinetic fitting with fixed global Rmax. d, The highest affinity HA binder, HA_20, binds with a KD of 28 nM. c,d, Yellow or orange, target or hotspot residues; grey, design model; purple, AF2 prediction (r.m.s.d. AF2 versus design). Binders: IL7Ra_55 (2.1 Å), InsulinR_30 (2.6 Å), PDL1_77 (1.5 Å), TrkA_88 (1.4 Å) (left to right in c) and HA_20 (1.7 Å) (d). e, Cryo-EM 2D class averages of HA_20 bound to influenza HA, strain A/USA:Iowa/1943 H1N1 (scale bar, 10 nm). f, 2.9 Å cryo-EM 3D reconstruction of the complex viewed along two orthogonal axes. HA_20 (purple) is bound to H1 along the stem of all three subunits. g, The cryo-EM structure of the HA_20 binder in complex closely matches the design model (r.m.s.d. to RFdiffusion design, 0.63 Å; yellow, influenza HA). h, Structure of the HA_20 binder alone superimposed on the design model viewed along two orthogonal axes. For cryo-EM panels, yellow, Influenza H1 map and/or structure; grey, HA_20 binder design model; purple, HA_20 binder map or structure.
a,RFdiffusion在给定界面热点残基的靶标和规格的情况下生成蛋白质结合剂。b, 从头结合剂设计为5个蛋白质靶标;所有靶标均鉴定出甲型流感 H1 HA、IL-7Rα、InsR、PD-L1 和 TrkA 以及 BLI 反应大于或等于阳性对照的 50% 的命中。对于 IL-7Rα、InsR、PD-L1 和 TrkA,RFdiffusion 的成功率比原始设计活动高出大约两个数量级。我们将一个数量级归因于 RFdiffusion,第二个数量级归因于 AF2 滤波(如果使用 AF2 滤波,则先前活动的估计成功率:HA,0%;IL-7Rα,2.2%;InsR,5.5%;PD-L1,3.7%;TrkA,1.5%)。c,对于IL-7Rα、InsR、PD-L1和TrkA,最高亲和力的结合剂如BLI滴定系列所示。报告的 K D 值基于具有固定全局 R max 的全局动力学拟合。d, 最高亲和力的HA结合剂HA_20,结合的K D 为28 nM。c,d: 黄色或橙色,靶斑或热点残留;灰色,设计模型;紫色,AF2 预测(r.m.s.d. AF2 与设计)。粘合剂:IL7Ra_55 (2.1 Å)、InsulinR_30 (2.6 Å)、PDL1_77 (1.5 Å)、TrkA_88 (1.4 Å)(c中从左到右)和 HA_20 (1.7 Å) (d)。e,与流感 HA 结合的 HA_20 的冷冻电镜 2D 类平均值,菌株 A/USA:Iowa/1943 H1N1(比例尺,10 nm)。f, 2.9 Å 冷冻电镜 3D 重建沿两个正交轴观察的复合物。HA_20(紫色)沿着所有三个亚基的茎与 H1 结合。g, 复合物中HA_20粘合剂的冷冻电镜结构与设计模型非常吻合(r.m.s.d.到RFdiffusion设计,0.63 Å;黄色,流感HA)。h,单独叠加在沿两个正交轴查看的设计模型上的HA_20粘结剂结构。 对于冷冻电镜面板,黄色,H1流感图谱和/或结构;灰色、HA_20粘结剂设计模型;紫色,HA_20粘合剂映射或结构。
To compare RFdiffusion to previous binder design methods, we performed binder design campaigns against five targets: Influenza A H1 Haemagglutinin (HA)52, Interleukin-7 Receptor-ɑ (IL-7Rɑ)12, Programmed Death-Ligand 1 (PD-L1)12, Insulin Receptor (InsR) and Tropomyosin Receptor Kinase A (TrkA)12. We designed putative binders to each target, both with and without conditioning on compatible fold information, with high in silico success rates (Extended Data Fig. 8e,f). Designs were filtered by AF2 confidence in the interface and monomer structure26, and 95 were selected for each target for experimental characterization.
为了将RFdiffusion与以前的粘合剂设计方法进行比较,我们针对五个靶点进行了粘合剂设计活动:甲型流感H1血凝素(HA)、 52 白细胞介素-7受体-ɑ(IL-7Rɑ)、 12 程序性死亡配体1(PD-L1)、 12 胰岛素受体(InsR)和原肌球蛋白受体激酶A(TrkA)。 12 我们针对每个靶标设计了假定的粘合剂,无论是否对兼容的折叠信息进行调节,计算机模拟成功率都很高(扩展数据图8e,f)。通过界面和单体结构 26 的AF2置信度过滤设计,每个靶标选择95个进行实验表征。
The designed binders were expressed in E. coli and purified, and binding was assessed through single point biolayer interferometry (BLI) screening at 10 μM binder concentration (Extended Data Fig. 8g). The overall experimental success rate, defined as binding at or above 50% of the maximal response for the positive control, was 19% (this is a conservative estimate as some designs that showed binding had insufficient material to permit screening at 10 μM: Extended Data Fig. 8g); an increase of roughly two orders of magnitude over our previous Rosetta-based method on the same targets (Fig. 6b). Binders were identified for all five targets, with fewer than 100 designs tested per target compared to thousands in previous studies. Full BLI titrations for a subset of the designs showed nanomolar affinities with no further experimental optimization, including HA and IL-7Rɑ binders with affinities of roughly 30 nM (Fig. 6c). Binding interfaces were often highly distinct from interfaces to these targets in the PDB (Supplementary Figs. 11 and 12). To assess binder specificity, six of the highest affinity IL-7Rɑ binders were assessed by means of competition BLI, and all six competed for binding with a structurally validated positive control binding to the same site (Supplementary Fig. 10a; further work is required to fully characterize proteome-wide specificity).
将设计的结合剂在大肠杆菌中表达并纯化,并通过10μM结合剂浓度的单点生物层干涉测量(BLI)筛选评估结合(扩展数据图8g)。总体实验成功率(定义为结合率达到或高于阳性对照最大反应的 50%)为 19%(这是一个保守估计,因为一些显示结合的设计没有足够的材料允许在 10 μM 下进行筛选:扩展数据图 8g);比我们之前在相同目标上基于罗塞塔的方法增加了大约两个数量级(图6b)。所有五个靶点都确定了粘合剂,每个靶标测试的设计少于100个,而以前的研究中有数千个。部分设计的全BLI滴定显示纳摩尔亲和力,无需进一步的实验优化,包括亲和力约为30 nM的HA和IL-7Rɑ结合剂(图6c)。在PDB中,结合界面通常与这些靶标的界面高度不同(补充图11和12)。为了评估结合剂特异性,通过竞争BLI评估了六种亲和力最高的IL-7Rɑ结合剂,并且所有六种结合剂都与结构验证的阳性对照结合到同一位点(补充图10a;需要进一步的工作来充分表征蛋白质组范围的特异性)。
We solved the structure of the highest affinity Influenza binder, HA_20, in complex with Iowa43 HA using cryo-EM (Extended Data Table 1). Raw electron micrographs revealed a well-folded HA glycoprotein with clearly discernible side, top and tilted view orientations suspended in a thin layer of vitreous ice (Extended Data Fig. 9a). The 2D class averages further show clear secondary structure elements corresponding to both Iowa43 HA (Extended Data Fig. 9b), as well as the HA_20 binder bound to the stem (Fig 6e). The 3D heterogenous refinement without symmetry revealed full occupancy of all three HA stem epitopes by the HA_20 binder. A final non-uniform 3D refinement reconstruction with C3 symmetry yielded a 2.9 Å map of the HA/HA_20 protein–protein complex (Fig 6f) and corresponding 3D structure that almost perfectly matches the computational design model (0.63 Å, Fig 6f,g; the sidechain interactions at the interface are very different from the closest structure in the PDB; Extended Data Fig. 9h). Over the binder alone, the experimental structure deviates from the RFdiffusion design by only 0.6 Å (Fig. 6h). These results demonstrate the ability of RFdiffusion to generate new proteins with atomic level accuracy, and to precisely target functionally relevant sites on therapeutically important proteins.
我们使用冷冻电镜解决了与 Iowa43 HA 复合物中最高亲和力的流感结合剂 HA_20 的结构(扩展数据表 1)。原始电子显微照片显示,折叠良好的HA糖蛋白具有清晰可辨的侧面、顶部和倾斜视图方向,悬浮在一层薄薄的玻璃冰中(扩展数据图9a)。二维类平均值进一步显示了对应于 Iowa43 HA 的清晰二级结构元素(扩展数据图 9b)以及与茎结合的HA_20粘合剂(图 6e)。无对称性的3D异质细化揭示了HA_20结合剂完全占据了所有三个HA茎表位。最终,具有C3对称性的非均匀3D细化重建产生了HA/HA_20蛋白-蛋白质复合物的2.9 Å图谱(图6f)和相应的3D结构图谱,该图几乎与计算设计模型完美匹配(0.63 Å,图6f,g;界面处的侧链相互作用与PDB中最接近的结构有很大不同;扩展数据图9h)。仅就粘结剂而言,实验结构仅与RFdiffusion设计相差0.6 Å(图6h)。这些结果证明了RFdiffusion能够以原子级精度生成新蛋白质,并精确靶向治疗重要蛋白质上的功能相关位点。
Discussion 讨论
RFdiffusion is a comprehensive improvement over current protein design methods. RFdiffusion readily generates diverse unconditional designs up to 600 residues in length that are accurately predicted by AF2, far exceeding the complexity and accuracy achieved by most previous methods (a recent Hallucination-based approach also achieved high unconditional performance53). Half of our tested unconditional designs express in a soluble way, and have circular dichroism spectra consistent with the design models and high thermostability. Despite their substantially increased complexity, the ideality and stability of RFdiffusion designs is akin to that of de novo protein designs generated using previous methods such as Rosetta. RFdiffusion enables generation of higher-order architectures with any desired symmetry, unlike Hallucination methods, which have so far been limited to cyclic symmetries. Electron microscopy confirmed that the structures of these oligomers are very similar to the design models, which in many cases show little global similarity to known protein oligomers.
RFdiffusion是对当前蛋白质设计方法的全面改进。RFdiffusion 很容易生成各种无条件设计,长度可达 600 个残基,这些设计可通过 AF2 准确预测,远远超过大多数先前方法所达到的复杂性和准确性(最近一种基于幻觉的方法也实现了高无条件性能 53 )。我们测试的无条件设计中有一半以可溶方式表达,并具有与设计模型一致的圆二色光谱和高热稳定性。尽管它们的复杂性大大增加,但RF扩散设计的理想性和稳定性类似于使用先前方法(如Rosetta)生成的从头蛋白质设计。RFdiffusion能够生成具有任何所需对称性的高阶架构,这与幻觉方法不同,幻觉方法迄今为止仅限于循环对称性。电子显微镜证实,这些低聚物的结构与设计模型非常相似,在许多情况下,设计模型与已知的蛋白质低聚物几乎没有全局相似性。
There has been recent progress in scaffolding protein functional motifs using deep-learning methods (RF Hallucination, RFjoint Inpainting and diffusion), but Hallucination is slow for large systems, Inpainting fails when insufficient starting information is provided and previous diffusion methods had low accuracy. RFdiffusion outperforms these previous methods in the complexity of the motifs that can be scaffolded, the precision with which sidechains are positioned (for catalysis and other functions), and the accuracy of motif recapitulation by AF2. The design of MDM2 binding proteins with three orders of magnitude higher affinities than the scaffolded P53 motif demonstrates the robustness of RFdiffusion motif scaffolding. Combining accurate motif scaffolding with the design of symmetric assemblies enabled consistent and atomically precise positioning of sidechains to coordinate Ni2+ ions across diverse tetrameric assemblies
最近在使用深度学习方法(RF 幻觉、RF joint Inpainting 和 diffusion)构建蛋白质功能基序方面取得了进展,但对于大型系统来说,幻觉速度很慢,当提供起始信息不足时,Inpainting 会失败,并且以前的扩散方法精度较低。RFdiffusion在可以搭建的基序的复杂性、侧链定位的精度(用于催化和其他功能)以及AF2对基序概括的准确性方面优于以前的这些方法。MDM2结合蛋白的设计比支架的P53基序具有三个数量级的亲和力,证明了RFdiffusion基序支架的稳健性。将精确的基序支架与对称组件的设计相结合,可以实现侧链的一致和原子精确定位,以协调不同四聚体组件中的 2+ 镍离子
For binder design from target structural information alone, previous work required testing tens of thousands of sequences12. RFdiffusion, when combined with improved filtering26 raises experimental success rates by two orders of magnitude; high-affinity binders can be identified from dozens of designs, in many cases eliminating the requirement for slow and expensive high-throughput screening (at least for the non-polar sites targeted here; further studies will be required to assess success rates on more polar target sites and sites without native binding partners). A high-resolution cryo-EM structure of one of these designs in complex with influenza HA shows that RFdiffusion can design functional proteins with atomic accuracy. Vázquez Torres et al. demonstrate the ability of RFdiffusion to design picomolar affinity binders to flexible helical peptides54, further highlighting its use for de novo binder design. Vázquez Torres et al. also show how RFdiffusion can be extended for protein model refinement by partial noising and denoising, which enables tuneable sampling around a given input structure. For peptide binder design, this enabled increases in affinity of nearly three orders of magnitude without high-throughput screening.
仅根据目标结构信息进行粘结剂设计,以前的工作需要测试数以万计的序列 12 。RFdiffusion与改进的 26 滤波相结合,将实验成功率提高了两个数量级;可以从数十种设计中鉴定出高亲和力结合剂,在许多情况下,无需进行缓慢而昂贵的高通量筛选(至少对于此处针对的非极性位点;需要进一步的研究来评估更多极性靶位点和没有天然结合伴侣的位点的成功率)。其中一种设计与流感 HA 复合物的高分辨率冷冻电镜结构表明,RFdiffusion 可以设计具有原子精度的功能蛋白。Vázquez Torres等人证明了RFdiffusion能够将皮摩尔亲和力结合剂设计成柔性螺旋肽 54 ,进一步突出了它在从头粘合剂设计中的应用。Vázquez Torres 等人还展示了如何通过部分噪声和去噪来扩展 RFdiffusion 以改进蛋白质模型,从而可以围绕给定的输入结构进行可调采样。对于肽结合剂设计,无需高通量筛选即可将亲和力提高近三个数量级。
The breadth and complexity of problems solvable with RFdiffusion and the robustness and accuracy of the solutions far exceeds what has been achieved previously. In a manner reminiscent of the generation of images from text prompts, RFdiffusion makes possible, with minimal specialist knowledge, the generation of functional proteins from minimal molecular specifications (for example, high-affinity binders to a user-specified target protein, and diverse protein assemblies from user-specified symmetries).
RFdiffusion可解决的问题的广度和复杂性以及解决方案的鲁棒性和准确性远远超过了以前所取得的成就。RFdiffusion以一种让人联想到从文本提示生成图像的方式,以最少的专业知识,从最小的分子规格(例如,用户指定的靶蛋白的高亲和力结合剂,以及来自用户指定的对称性的各种蛋白质组装)生成功能蛋白。
The power and scope of RFdiffusion can be extended in several directions. RF has recently been extended to nucleic acids and protein–nucleic acid complexes55, which should enable RFdiffusion to design nucleic acid binding proteins and perhaps folded RNA structures. Extension of RF to incorporate ligands should similarly enable extension of RFdiffusion to explicitly model ligand atoms, and allow the design of protein–ligand interactions. The ability to customize RFdiffusion to specific design challenges by addition of external potentials and by fine-tuning (as illustrated here for catalytic site scaffolding, binder-targeting and fold specification), along with continued improvements to the underlying methodology, should enable de novo protein design to achieve still higher levels of complexity, to approach and, in some cases, surpass what natural evolution has achieved.
RFdiffusion 的功率和范围可以在多个方向上扩展。RF最近被扩展到核酸和蛋白质-核酸复合物 55 ,这应该使RFdiffusion能够设计核酸结合蛋白,并可能设计折叠的RNA结构。RF的扩展以合并配体应该同样能够扩展RFdiffusion以显式模拟配体原子,并允许设计蛋白质-配体相互作用。通过添加外部电位和微调(如此处所示的催化位点支架、粘合剂靶向和折叠规范)来定制 RFdiffusion 以应对特定设计挑战的能力,以及对基础方法的持续改进,应该使从头蛋白质设计能够实现更高水平的复杂性,接近并在某些情况下超越自然进化所取得的成就。
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
有关研究设计的更多信息,请参阅本文链接的《自然投资组合报告摘要》。
Data availability
Design structures, AF2 models and experimental measurements are available at https://figshare.com/s/439fdd59488215753bc3. Cryo-EM maps and corresponding atomic models for the Influenza HA binder in Fig. 6d–h have been deposited in the PDB and the Electron Microscopy Data Bank under accession codes 8SK7 and EMDB-40557, respectively. Electron microscopy data collected for the HE0537 oligomer are available at EMDB-40602.
Code availability 代码可用性
Code for running RFdiffusion has been released on GitHub, free for academic, personal and commercial use at https://github.com/RosettaCommons/RFdiffusion. It is also available as a Google Colab notebook, accessible through GitHub.
运行 RFdiffusion 的代码已在 GitHub 上发布,可在 https://github.com/RosettaCommons/RFdiffusion 免费用于学术、个人和商业用途。它也可以作为 Google Colab 笔记本使用,可通过 GitHub 访问。
References 引用
Dauparas, J. et al. Robust deep learning-based protein sequence design using ProteinMPNN. Science 378, 49–56 (2022).
Dauparas, J. 等人。使用 ProteinMPNN 进行基于深度学习的稳健蛋白质序列设计。科学 378, 49–56 (2022)。Ferruz, N., Schmidt, S. & Höcker, B. ProtGPT2 is a deep unsupervised language model for protein design. Nat. Commun. 13, 4348 (2022).
Ferruz, N., Schmidt, S. & Höcker, B. ProtGPT2 是一种用于蛋白质设计的深度无监督语言模型。Nat. Commun.13, 4348 (2022).Singer, J. M. et al. Large-scale design and refinement of stable proteins using sequence-only models. PLoS ONE 17, e0265020 (2022).
Singer, J. M. 等人。使用纯序列模型对稳定蛋白进行大规模设计和改进。公共科学图书馆一号 17,e0265020 (2022)。Wang, J. et al. Scaffolding protein functional sites using deep learning. Science 377, 387–394 (2022).
Wang, J. 等人。使用深度学习搭建蛋白质功能位点。科学 377, 387–394 (2022)。Trippe, B. L. et al. Diffusion probabilistic modeling of protein backbones in 3D for the motif-scaffolding problem. in The Eleventh International Conference on Learning Representations (2023).
Trippe, B. L. 等人。蛋白质骨架的 3D 扩散概率建模,用于基序-支架问题。在第十一届学习表征国际会议(2023 年)中。Anishchenko, I. et al. De novo protein design by deep network hallucination. Nature 600, 547–552 (2021).
Anishchenko, I. 等人。通过深度网络幻觉进行从头蛋白质设计。自然 600, 547–552 (2021)。Wicky, B. I. M. et al. Hallucinating symmetric protein assemblies. Science 378, 56–61 (2022).
Wicky, B. I. M. 等人。幻觉对称蛋白质组装体。科学 378, 56–61 (2022)。Anand, N. & Achim, T. Protein structure and sequence generation with equivariant denoising diffusion probabilistic models. Preprint at https://doi.org/10.48550/arXiv.2205.15019 (2022).
Anand, N. & Achim, T. 使用等变去噪扩散概率模型生成蛋白质结构和序列。https://doi.org/10.48550/arXiv.2205.15019 (2022) 的预印本。Luo, S. et al. Antigen-specific antibody design and optimization with diffusion-based generative models. in Adv. Neural Information Processing Systems Vol. 35 (eds Koyejo, S. et al) 9754–9767 (Curran Associates, 2022).
Luo, S. 等人。使用基于扩散的生成模型进行抗原特异性抗体设计和优化。神经信息处理系统第 35 卷(Koyejo 等人编辑)9754–9767(Curran Associates,2022 年)。Sohl-Dickstein, J., Weiss, E. A., Maheswaranathan, N. & Ganguli, S. Deep unsupervised learning using nonequilibrium thermodynamics. in Proc. 32nd International Conference on Machine Learning Vol. 37 (eds Bach, Francis and Blei, David) 2256–2265 (PMLR, 2015).
Sohl-Dickstein, J., Weiss, E. A., Maheswaranathan, N. & Ganguli, S. 使用非平衡热力学的深度无监督学习。第 32 届机器学习国际会议第 37 卷(Bach、Francis 和 Blei、David 编辑)2256–2265(PMLR,2015 年)。Ho, J., Jain, A. & Abbeel, P. Denoising diffusion probabilistic models. in Adv. Neural Information Processing Systems Vol. 33 (eds Larochelle, H. et al.) 6840–6851 (Curran Associates, 2020).
Ho, J., Jain, A. & Abbeel, P. 去噪扩散概率模型。神经信息处理系统第 33 卷(Larochelle, H. 等人编辑)6840–6851(Curran Associates,2020 年)。Cao, L. et al. Design of protein-binding proteins from the target structure alone. Nature 605, 551–560 (2022).
Cao, L. et al.仅从靶标结构设计蛋白质结合蛋白。自然 605, 551–560 (2022)。Kuhlman, B. et al. Design of a novel globular protein fold with atomic-level accuracy. Science 302, 1364–1368 (2003).
Kuhlman, B. 等人。具有原子级精度的新型球状蛋白质折叠设计。科学 302, 1364–1368 (2003)。Ramesh, A. et al. Zero-shot text-to-image generation. in Proc. 38th International Conference on Machine Learning Vol. 139 (eds Meila, M. & Zhang, T.) 8821–8831 (PMLR, 2021).
Ramesh, A. 等人。零样本文本到图像生成。第 38 届机器学习国际会议第 139 卷(编辑 Meila, M. & Zhang, T.) 8821–8831 (PMLR, 2021)。Saharia, C. et al. Photorealistic text-to-image diffusion models with deep language understanding. in Adv. Neural Information Processing Systems Vol. 35 (eds Koyejo, S. et al) 36479–36494 (Curran Associates, 2022).
Saharia, C. 等人。具有深刻语言理解的逼真的文本到图像扩散模型。神经信息处理系统第 35 卷(编辑 Koyejo, S. 等人)36479–36494(Curran Associates,2022 年)。Wu, K. E. et al. Protein structure generation via folding diffusion. Preprint at https://doi.org/10.48550/arXiv.2209.15611 (2022).
Wu, K. E. 等人。通过折叠扩散产生蛋白质结构。预印本 https://doi.org/10.48550/arXiv.2209.15611(2022 年)。Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Jumper, J. 等人。使用 AlphaFold 进行高度准确的蛋白质结构预测。自然 596, 583–589 (2021)。Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021).
Baek, M. 等人。使用三轨神经网络准确预测蛋白质结构和相互作用。科学 373, 871–876 (2021)。Watson, J. L., Bera, A., Juergens, D., Wang, J. & Baker, D. X-ray crystallographic validation of design from this paper. Science 377, 387–394 (2022).
Watson, J. L., Bera, A., Juergens, D., Wang, J. & Baker, D. 本文设计的X射线晶体学验证。科学 377, 387–394 (2022)。Wu, R. et al. High-resolution de novo structure prediction from primary sequence. Preprint at https://doi.org/10.1101/2022.07.21.500999 (2022).
Wu, R. 等人。从初级序列进行高分辨率从头结构预测。https://doi.org/10.1101/2022.07.21.500999 (2022) 的预印本。Lin, Z. et al. Language models of protein sequences at the scale of evolution enable accurate structure prediction. Science 379, 1123–1130 (2023).
Lin, Z. 等人。进化尺度上的蛋白质序列语言模型能够实现准确的结构预测。科学 379, 1123–1130 (2023)。Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Berman, HM 等人。蛋白质数据库。核酸研究 28, 235–242 (2000)。De Bortoli, V. et al. Riemannian score-based generative modelling. in Adv. Neural Information Processing Systems Vol. 35 (eds Koyejo, S. et al) 2406–2422 (Curran Associates, 2022).
De Bortoli, V. 等人。基于黎曼分数的生成建模。在 Adv. Neural Information Processing Systems Vol. 35 (eds Koyejo, S. et al) 2406–2422 (Curran Associates, 2022) 中。Leach, A., Schmon, S. M., Degiacomi, M. T. & Willcocks, C. G. Denoising diffusion probabilistic models on SO(3) for rotational alignment. In Proc. ICLR 2022 Workshop on Geometrical and Topological Representation Learning (2022).
Leach, A., Schmon, S. M., Degiacomi, M. T. & Willcocks, C. G. SO(3) 上的去噪扩散概率模型用于旋转对准。ICLR 2022 几何和拓扑表示学习研讨会 (2022)。Chen, T., Zhang, R. & Hinton, G. Analog bits: generating discrete data using diffusion models with self-conditioning. in The Eleventh International Conference on Learning Representations (2023).
Chen, T., Zhang, R. & Hinton, G. 模拟位:使用具有自调节功能的扩散模型生成离散数据。在第十一届学习表征国际会议(2023 年)中。Bennett, N.R. et al. Improving de novo protein binder design with deep learning. Nat. Commun. 14, 2625 (2023).
Bennett, N.R. 等人。通过深度学习改进从头蛋白质结合剂设计。Nat. Commun.14, 2625 (2023).Anand, N. & Huang, P. Generative modeling for protein structures. in Adv. Neural Information Processing Systems Vol. 31 (eds Bengio, S. et al) (Curran Associates, 2018).
Anand, N. & Huang, P. 蛋白质结构的生成建模。神经信息处理系统第 31 卷(Bengio, S. 等人编辑)(Curran Associates,2018 年)。Ingraham, J. et al. Illuminating protein space with a programmable generative model. Preprint at bioRxiv https://doi.org/10.1101/2022.12.01.518682 (2022).
英格拉汉姆,J.等人。使用可编程生成模型照亮蛋白质空间。bioRxiv https://doi.org/10.1101/2022.12.01.518682 (2022) 的预印本。Lee, J. S. & Kim, P. M. ProteinSGM: Score-based generative modeling for de novo protein design. Preprint at bioRxiv https://doi.org/10.1101/2022.07.13.499967 (2022).
Lee, J. S. & Kim, P. M. ProteinSGM:基于分数的从头蛋白质设计的生成建模。bioRxiv https://doi.org/10.1101/2022.07.13.499967 (2022) 的预印本。Onuchic, J. N., Luthey-Schulten, Z. & Wolynes, P. G. Theory of protein folding: the energy landscape perspective. Annu. Rev. Phys. Chem. 48, 545–600 (1997).
Onuchic, J. N., Luthey-Schulten, Z. & Wolynes, P. G. 蛋白质折叠理论:能源景观视角。年。Rev. Phys. Chem. 48, 545–600 (1997).Jendrusch, M., Korbel, J. O. & Sadiq, S. K. AlphaDesign: a de novo protein design framework based on AlphaFold. Preprint at bioRxiv https://doi.org/10.1101/2021.10.11.463937 (2021).
Jendrusch, M., Korbel, J. O. & Sadiq, S. K. AlphaDesign:基于AlphaFold的从头蛋白质设计框架。bioRxiv https://doi.org/10.1101/2021.10.11.463937 (2021) 的预印本。Basanta, B. et al. An enumerative algorithm for de novo design of proteins with diverse pocket structures. Proc. Natl Acad. Sci. USA 117, 22135–22145 (2020).
Basanta, B. 等人。一种用于从头设计具有不同口袋结构的蛋白质的枚举算法。美国国家科学院院刊 117, 22135–22145 (2020)。Pan, X. et al. Expanding the space of protein geometries by computational design of de novo fold families. Science 369, 1132–1136 (2020).
Pan, X. 等人。通过从头折叠家族的计算设计来扩展蛋白质几何空间。科学 369, 1132–1136 (2020)。Marcandalli, J. et al. Induction of potent neutralizing antibody responses by a designed protein nanoparticle vaccine for respiratory syncytial virus. Cell 176, 1420–1431.e17 (2019).
Marcandalli, J. 等人。通过设计的用于呼吸道合胞病毒的蛋白质纳米颗粒疫苗诱导有效的中和抗体反应。单元格 176, 1420–1431.e17 (2019)。Butterfield, G. L. et al. Evolution of a designed protein assembly encapsulating its own RNA genome. Nature 552, 415–420 (2017).
Butterfield, G. L. 等人。设计蛋白质组装的进化,封装其自身的RNA基因组。自然 552, 415–420 (2017)。Goodsell, D. S. & Olson, A. J. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 29, 105–153 (2000).
Goodsell, D. S. & Olson, A. J. 结构对称性和蛋白质功能。年。生物物理学牧师。生物摩尔。结构。29, 105–153 (2000).Sterner, R. & Höcker, B. Catalytic versatility, stability, and evolution of the (βα)8-barrel enzyme fold. Chem. Rev. 105, 4038–4055 (2005).
Sterner, R. & Höcker, B. (βα) 8 -桶状酶折叠的催化多功能性、稳定性和进化。Chem. Rev. 105, 4038–4055 (2005).Sesterhenn, F. et al. De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science 368, eaay5051 (2020).
Sesterhenn, F. 等人。从头蛋白质设计可精确诱导 RSV 中和抗体。科学 368, eaay5051 (2020).Yang, C. et al. Bottom-up de novo design of functional proteins with complex structural features. Nat. Chem. Biol. 17, 492–500 (2021).
Yang, C. 等人。具有复杂结构特征的功能蛋白的自下而上的从头设计。Nat. Chem. Biol. 17, 492–500 (2021).Glasgow, A. et al. Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc. Natl Acad. Sci. USA 117, 28046–28055 (2020).
格拉斯哥,A.等人。工程化的 ACE2 受体陷阱可有效中和 SARS-CoV-2。美国国家科学院院刊 117, 28046–28055 (2020)。Chène, P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat. Rev. Cancer 3, 102–109 (2003).
抑制p53-MDM2相互作用:癌症治疗的重要靶点。Nat. Rev. 癌症 3, 102–109 (2003)。Kussie, P. H. et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274, 948–953 (1996).
Kussie, P. H. 等人。与 p53 抑癌基因反式激活结构域结合的 MDM2 癌蛋白的结构。科学 274, 948–953 (1996)。Hunt, A. C. et al. Multivalent designed proteins neutralize SARS-CoV-2 variants of concern and confer protection against infection in mice. Sci. Transl. Med. 14, eabn1252 (2022).
Hunt, A. C. 等人。多价设计的蛋白质可中和值得关注的 SARS-CoV-2 变体,并保护小鼠免受感染。Sci. Transl. Med. 14, eabn1252 (2022).Silverman, J. et al. Multivalent avimer proteins evolved by exon shuffling of a family of human receptor domains. Nat. Biotechnol. 23, 1556–1561 (2005).
西尔弗曼,J.等人。多价 avimer 蛋白通过人类受体结构域家族的外显子洗牌进化而来。国家生物技术。23, 1556–1561 (2005).Detalle, L. et al. Generation and characterization of ALX-0171, a potent novel therapeutic nanobody for the treatment of respiratory syncytial virus infection. Antimicrob. Agents Chemother. 60, 6–13 (2016).
Detalle, L. 等人。ALX-0171的生成和表征,ALX-0171是一种用于治疗呼吸道合胞病毒感染的强效新型治疗性纳米抗体。抗菌剂。特工Chemother。60, 6–13 (2016).Strauch, E.-M. et al. Computational design of trimeric influenza-neutralizing proteins targeting the hemagglutinin receptor binding site. Nat. Biotechnol. 35, 667–671 (2017).
施特劳赫,EM等。靶向血凝素受体结合位点的三聚体流感中和蛋白的计算设计。国家生物技术。35, 667–671 (2017).Boyoglu-Barnum, S. et al. Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature 592, 623–628 (2021).
Boyoglu-Barnum, S. 等人。四价流感纳米颗粒疫苗可诱导广泛的保护作用。自然 592, 623–628 (2021)。Walls, A. C. et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell 183, 1367–1382.e17 (2020).
Walls, A. C. 等人。通过设计的 SARS-CoV-2 蛋白纳米颗粒疫苗引发有效的中和抗体反应。单元格 183, 1367–1382.e17 (2020)。Salgado, E. N., Lewis, R. A., Mossin, S., Rheingold, A. L. & Tezcan, F. A. Control of protein oligomerization symmetry by metal coordination: C2 and C3 symmetrical assemblies through CuII and NiII coordination. Inorg. Chem. 48, 2726–2728 (2009).
Salgado, E. N., Lewis, R. A., Mossin, S., Rheingold, A. L. & Tezcan, F. A. 通过金属配位控制蛋白质寡聚对称性:通过 Cu II 和 Ni II 配位控制 C 2 和 C 3 对称组装。伊诺格。Chem. 48, 2726–2728 (2009).Salgado, E. N. et al. Metal templated design of protein interfaces. Proc. Natl Acad. Sci. USA 107, 1827–1832 (2010).
Salgado, E. N. 等人。蛋白质界面的金属模板化设计。美国国家科学院院刊 107, 1827–1832 (2010)。Quijano-Rubio, A., Ulge, U. Y., Walkey, C. D. & Silva, D.-A. The advent of de novo proteins for cancer immunotherapy. Curr. Opin. Chem. Biol. 56, 119–128 (2020).
Quijano-Rubio, A., Ulge, U. Y., Walkey, C. D. & Silva, D.-A.用于癌症免疫治疗的从头蛋白的出现。卷曲。意见。化学生物学 56, 119–128 (2020)。Chevalier, A. et al. Massively parallel de novo protein design for targeted therapeutics. Nature 550, 74–79 (2017).
Chevalier, A. 等人。用于靶向治疗的大规模并行从头蛋白质设计。自然 550, 74–79 (2017)。Frank, C. et al. Efficient and scalable de novo protein design using a relaxed sequence space. Preprint at bioRxiv https://doi.org/10.1101/2023.02.24.529906 (2023).
弗兰克,C.等人。使用宽松的序列空间进行高效且可扩展的从头蛋白质设计。bioRxiv https://doi.org/10.1101/2023.02.24.529906 (2023) 的预印本。Torres, S. V. et al. De novo design of high-affinity protein binders to bioactive helical peptides. Preprint at bioRxiv https://doi.org/10.1101/2022.12.10.519862 (2022).
Torres, S. V. 等人。从头设计生物活性螺旋肽的高亲和力蛋白质结合剂。bioRxiv https://doi.org/10.1101/2022.12.10.519862 (2022) 的预印本。Baek, M., McHugh, R., Anishchenko, I., Baker, D. & DiMaio, F. Accurate prediction of nucleic acid and protein-nucleic acid complexes using RoseTTAFoldNA. Preprint at bioRxiv https://doi.org/10.1101/2022.09.09.507333 (2022).
Baek, M., McHugh, R., Anishchenko, I., Baker, D. & DiMaio, F. 使用 RoseTTAFoldNA 准确预测核酸和蛋白质-核酸复合物。bioRxiv https://doi.org/10.1101/2022.09.09.507333 (2022) 的预印本。Yeh, A. H.-W. et al. De novo design of luciferases using deep learning. Nature 614, 774–780 (2023).
叶,A.H.-W.等。使用深度学习对荧光素酶进行从头设计。自然 614, 774–780 (2023)。Ribeiro, A. J. M. et al. Mechanism and Catalytic Site Atlas (M-CSA): a database of enzyme reaction mechanisms and active sites. Nucleic Acids Res. 46, D618–D623 (2018).
Ribeiro, A. J. M. 等人。机理和催化位点图谱(M-CSA):酶反应机理和活性位点的数据库。核酸研究 46, D618–D623 (2018)。Leaver-Fay, A. et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574 (2011).
Leaver-Fay, A. et al. ROSETTA3:用于模拟和设计大分子的面向对象的软件套件。方法:Enzymol。487, 545–574 (2011).
Acknowledgements 确认
We thank N. Anand and D. Tischer for helpful discussions, and I. Kalvet and Y. Kipnis for providing helpful Rosetta scripts. We thank A. Dosey for the provision of purified influenza HA protein. We thank R. Wu, J. Mou, K. Choi, L. Wu and D. Blei for valuable feedback during writing. We thank I. Haydon for help with graphics. We also thank L. Goldschmidt and K. VanWormer, respectively, for maintaining the computational and wet laboratory resources at the Institute for Protein Design. This work was supported by gifts from Microsoft (D.J., M.B. and D.B.), Amgen (J.L.W.), the Audacious Project at the Institute for Protein Design (B.L.T., I.S., J.Y., H.E. and D.B.), the Washington State General Operating Fund supporting the Institute for Protein Design (P.V. and I.S.), grant no. INV-010680 from the Bill and Melinda Gates Foundation (W.B.A., D.J., J.W. and D.B.), grant no. DE-SC0018940 MOD03 from the US Department of Energy Office of Science (A.J.B. and D.B.), grant no. 5U19AG065156-02 from the National Institute for Aging (S.V.T. and D.B.), an EMBO long-term fellowship no. ALTF 139-2018 (B.I.M.W.), the Open Philanthropy Project Improving Protein Design Fund (R.J.R. and D.B.), The Donald and Jo Anne Petersen Endowment for Accelerating Advancements in Alzheimer’s Disease Research (N.R.B.), a Washington Research Foundation Fellowship (S.J.P.), a Human Frontier Science Program Cross Disciplinary Fellowship (grant no. LT000395/2020-C, L.F.M.), an EMBO Non-Stipendiary Fellowship (grant no. ALTF 1047-2019, L.F.M.), the Defense Threat Reduction Agency grant nos. HDTRA1-19-1-0003 (N.H. and D.B.) and HDTRA12210012 (F.D.), the Institute for Protein Design Breakthrough Fund (A.C. and D.B.), an EMBO Postdoctoral Fellowship (grant no. ALTF 292-2022, J.L.W.) and the Howard Hughes Medical Institute (A.C., W.S., R.J.R. and D.B.), an NSF-GRFP (J.Y.), an NSF Expeditions grant (no. 1918839, J.Y., R.B. and T.S.J.), the Machine Learning for Pharmaceutical Discovery and Synthesis consortium (J.Y., R.B. and T.S.J.), the Abdul Latif Jameel Clinic for Machine Learning in Health (J.Y., R.B. and T.S.J.), the DTRA Discovery of Medical Countermeasures Against New and Emerging threats program (J.Y., R.B. and T.S.J.), EPSRC Prosperity Partnership grant no. EP/T005386/1 (E.M.) and the DARPA Accelerated Molecular Discovery program and the Sanofi Computational Antibody Design grant (J.Y., R.B. and T.S.J.). We thank Microsoft and AWS for generous gifts of cloud computing resources.
我们感谢 N. Anand 和 D. Tischer 的有益讨论,感谢 I. Kalvet 和 Y. Kipnis 提供有用的 Rosetta 脚本。我们感谢 A. Dosey 提供纯化的流感 HA 蛋白。我们感谢 R. Wu、J. Mou、K. Choi、L. Wu 和 D. Blei 在写作过程中提供的宝贵反馈。我们感谢 I. Haydon 在图形方面的帮助。我们还分别感谢 L. Goldschmidt 和 K. VanWormer 维护蛋白质设计研究所的计算和湿实验室资源。这项工作得到了Microsoft(D.J.、M.B. 和 D.B.)、安进 (J.L.W.)、蛋白质设计研究所的大胆项目(B.L.T.、I.S.、J.Y.、H.E. 和 D.B.)、支持蛋白质设计研究所(P.V. 和 I.S.)的华盛顿州总运营基金的捐赠,资助编号。来自比尔和梅琳达·盖茨基金会(W.B.A.、D.J.、J.W. 和 D.B.)的 INV-010680,授权号。DE-SC0018940 MOD03 来自美国能源部科学办公室(AJB 和 DB),来自美国国家老龄化研究所(S.V.T. 和 D.B.)的资助号 5U19AG065156-02,EMBO 长期奖学金编号。ALTF 139-2018 (B.I.M.W.)、开放慈善项目改善蛋白质设计基金(RJR 和 DB)、Donald 和 Jo Anne Petersen 加速阿尔茨海默病研究进展基金会 (N.R.B.)、华盛顿研究基金会奖学金 (S.J.P.)、人类前沿科学计划跨学科奖学金(批准号。LT000395/2020-C, L.F.M.),EMBO非津贴奖学金(授予号。ALTF 1047-2019,L.F.M.),国防威胁减少局拨款编号。HDTRA1-19-1-0003(NH和DB)和HDTRA12210012(FD),蛋白质设计突破基金研究所(AC和DB),EMBO博士后奖学金(资助号。ALTF 292-2022,J.L.W.)和霍华德休斯医学研究所(A.C.、W.S.、R.J.R. 和 D.B.)、NSF-GRFP (J.Y.)、NSF Expeditions 赠款(编号 1918839、J.Y.、R.B. 和 T.S.J.)、用于药物发现和合成的机器学习联盟(J.Y.、R.B. 和 T.S.J.)、安利捷健康机器学习诊所(J.Y., R.B. 和 T.S.J.)、DTRA 发现针对新出现威胁的医疗对策计划(J.Y.、R.B. 和 T.S.J.),EPSRC 繁荣伙伴关系赠款编号。EP/T005386/1 (E.M.) 和 DARPA 加速分子发现计划以及赛诺菲计算抗体设计资助(J.Y.、R.B. 和 T.S.J.)。我们感谢 Microsoft 和 AWS 慷慨赠送的云计算资源。
Author information 作者信息
Authors and Affiliations
Contributions
J.L.W., D.J., N.R.B., B.L.T., J.Y. and D.B. conceived the study. J.L.W., D.J., N.R.B., W.A., B.L.T. and J.Y. trained RFdiffusion. B.L.T. and J.Y., with assistance from V.D.B. and E.M., extended diffusion to residue orientations. H.E.E., D.J., J.L.W., N.R.B., N.H., W.S., P.V. and I.S. generated experimentally characterized designs. W.A., B.L.T., J.Y., D.J., J.L.W. and N.R.B. generated computational designs. H.E.E., A.J.B., R.J.R., L.F.M., B.I.M.W., S.J.P., N.H., A.C., S.V.T., J.L.W. and B.L.T. experimentally characterized designs. J.W., A.L. and W.S. contributed additional code. S.O. implemented RFdiffusion on Google Colab. M.B. and F.D. trained RF. D.B., T.S.J. and R.B. offered supervision throughout the project. J.L.W., D.J., B.L.T., N.R.B., J.Y., H.E. and D.B. wrote the manuscript. All authors read and contributed to the manuscript. J.L.W. and D.J. agree that the order of their respective names may be changed for personal pursuits to best suit their own interests.
Corresponding author
Ethics declarations 道德宣言
Competing interests 利益争夺
The authors declare no competing interests.
作者声明没有竞争利益。
Peer review 同行审查
Peer review information 同行评议信息
Nature thanks Arne Elofsson, Giulia Palermo, Alex Pritzel and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
《自然》杂志感谢Arne Elofsson、Giulia Palermo、Alex Pritzel和其他匿名审稿人对这项工作的同行评审的贡献。
Additional information 其他信息:
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
出版商注:施普林格·自然(Springer Nature)对已出版地图和机构隶属关系中的管辖权主张保持中立。
Extended data figures and tables
扩展数据图表
Extended Data Fig. 1 Training ablations reveal determinants of RFdiffusion success.
扩展数据 图 1 训练消融揭示了射频扩散成功的决定因素。
A–C) RFdiffusion can generate high quality large unconditional monomers. Designs are routinely accurately recapitulated by AF2 (see also Fig. 2c), with high confidence (A) for proteins up to approximately 400 amino acids in length. B) Further orthogonal validation of designs by ESMFold. C) Recapitulation of the design structure is often better with ESMFold compared with AF2. For each backbone, the best of 8 ProteinMPNN sequences is plotted, with points therefore paired by backbone rather than sequence. D) Comparing RFdiffusion trained with MSE loss on Cα atoms and N-Cα-C backbone frames (Methods 2.5), rather than with FAPE loss8,17. The MSE loss is not invariant to the global coordinate frame, unlike FAPE loss, and is required for good performance at unconditional generation (left, two-proportion z-test of in silico success rate, n = 400 designs per condition, z = 4.1, p = 4.1e-5). For motif scaffolding problems, where the ‘motif’ provides a means to align the global coordinate frame between timesteps, FAPE loss performs approximately as well as MSE loss, suggesting the L2 nature of MSE loss (as opposed to the L1 loss in FAPE) is not empirically critical for performance. E) Allowing the model to condition on its X0 prediction at the previous timestep (see Supplementary Methods 2.4) improves designs. Designs with self-conditioning (pink) have improved recapitulation by AF2 (left) and better AF2 confidence in the prediction (right). Two-proportion z-test of in silico success rate, n = 800 designs per condition z = 11.4, p = 6.1e-30. F) RFdiffusion leverages the representations learned during RF pre-training. RFdiffusion fine-tuned from pre-trained RF (pink) comprehensively outperforms a model trained for an equivalent amount of time, from untrained weights (gray). For context, sequences generated by ProteinMPNN on these output backbones are little better than sampling ProteinMPNN sequences from random Gaussian-sampled coordinates (white). Two-proportion z-test of in silico success rate, pre-training vs without pre-training (or vs random noise; both have zero success rate), n = 800 designs per condition, z = 23.0, p = 3.1e-117. Note that the data in pink in D–F is the same data, reproduced in each plot for clarity. G) The median (by AF2 r.m.s.d. vs design) 300 amino acid unconditional sample highlighting the importance of self-conditioning and pre-training. Without pre-training (at least when trained with equivalent compute), RFdiffusion outputs bear little resemblance to proteins (gray, left). Without self-conditioning, outputs show characteristic protein secondary structures, but lack core-packing and ideality (gray, middle). With pre-training and self-conditioning, proteins are diverse and well-packed (pink, right). H) Greater coherence during unconditional denoising may partly explain the effect of self-conditioning. Successive X0 predictions are more similar when the model can self-condition (lower r.m.s.d. between X0 predictions, pink curve). Data are aggregated from unconditional design trajectories of 100, 200 and 300 residues. I) During the reverse (generation) process, the noise added at each step can be scaled (reduced). Reducing the noise scale improves the in silico design success rates (left, middle; two-proportion z-test of in silico success rate, n = 800 designs per condition, 0 vs 0.5: z = 1.7, p = 0.09, 0 vs 1: z = 6.5, p = 6.8e-11; 0.5 vs 1: z = 4.8, p = 1.4e-6). This comes at the expense of diversity, with the number of unique clusters at a TM-score cutoff of 0.6 reduced when noise is reduced (right). Note throughout this figure the 6EXZ_long benchmarking problem is abbreviated to 6EXZ for brevity. Boxplots represent median±IQR; tails: min/max excluding outliers (±1.5xIQR).
A–C) RF扩散可以生成高质量的大无条件单体。AF2通常可以精确地概括设计(另见图2c),对于长度高达约400个氨基酸的蛋白质具有高置信度(A)。B) 通过 ESMFold 对设计进行进一步的正交验证。C) 与 AF2 相比,ESMFold 对设计结构的概括通常更好。对于每个骨架,绘制了 8 个 ProteinMPNN 序列中最好的序列,因此按骨架而不是序列配对的点。D) 比较 Cα 原子和 N-Cα-C 骨架上 MSE 损失训练的 RF扩散(方法 2.5),而不是 FAPE 损失 8,17 。与FAPE损失不同,MSE损失与全局坐标系不变,并且是无条件生成时良好性能所必需的(左图,计算机成功率的双比例z检验,每个条件n = 400个设计,z = 4.1,p = 4.1e-5)。对于基序脚手架问题,其中“基序”提供了一种在时间步长之间对齐全局坐标系的方法,FAPE 损失的表现与 MSE 损失大致相同,这表明 MSE 损失的 L2 性质(与 FAPE 中的 L1 损失相反)在经验上对性能并不重要。E) 允许模型在前一个时间步长(参见补充方法 2.4)的 X 0 预测上进行条件调整,从而改进了设计。具有自调节功能(粉红色)的设计改进了 AF2 的概括(左)和更好的 AF2 预测置信度(右)。计算机成功率的双比例 z 检验,n = 800 个设计,每个条件 z = 11.4,p = 6.1e-30。F) RFdiffusion 利用了 RF 预训练期间学到的表示。 从预训练的 RF(粉红色)微调的 RFdiffusion 在综合上优于从未经训练的权重(灰色)训练了相同时间的模型。就上下文而言,ProteinMPNN 在这些输出主链上生成的序列比从随机高斯采样坐标(白色)采样 ProteinMPNN 序列好不了多少。计算机模拟成功率的双比例 z 检验,预训练与无预训练(或与随机噪声;两者的成功率为零),n = 每个条件 800 个设计,z = 23.0,p = 3.1e-117。请注意,D-F 中粉红色的数据是相同的数据,为清楚起见,在每个图中重现。G) 中位数(通过 AF2 r.m.s.d. 与设计)300 个氨基酸的无条件样本,突出了自我调节和预训练的重要性。如果没有预训练(至少在使用等效计算进行训练时),RFdiffusion 输出与蛋白质几乎没有相似之处(灰色,左)。如果没有自我调节,输出显示出特征性的蛋白质二级结构,但缺乏核心堆积和理想性(灰色,中间)。通过预训练和自我调节,蛋白质是多样化的,包装良好(粉红色,右)。H)无条件去噪期间更大的连贯性可以部分解释自我调节的效果。当模型可以自我调节时,连续的 X 0 预测更相似(X 0 预测之间的 r.m.s.d. 较低,粉红色曲线)。数据从 100、200 和 300 个残基的无条件设计轨迹中汇总而成。I) 在反向(生成)过程中,可以缩放(减少)每一步添加的噪声。降低噪声标度可提高计算机设计成功率(左,中;计算机成功率的双比例 z 检验,每个条件 n = 800 个设计,0 vs 0.5:z = 1.7,p = 0.09,0 vs 1:z = 6.5,p = 6.8e-11;0.5 对 1:z = 4.8,p = 1.4e-6)。这是以牺牲多样性为代价的,当噪声降低时,TM 分数截止值为 0.6 的独特集群数量会减少(右图)。请注意,为简洁起见,在本图中,6EXZ_long基准测试问题缩写为 6EXZ。箱线图表示中位数±IQR;尾部:最小值/最大值,不包括异常值 (±1.5xIQR)。
Extended Data Fig. 2 RFdiffusion learns the distribution of the denoising process, and inference efficiency can be improved.
扩展数据 图2 RFdiffusion学习去噪过程的分布,可以提高推理效率。
A) Analysis of simulated forward (noising) and reverse (denoising) trajectories shows that the distribution of Cα coordinates and residue orientations closely match, demonstrating that RFdiffusion has learned the distribution of the denoising process as desired. Left to right: i) average distance between a Cα coordinate at Xt and its position in X0; ii) average distance between a Cα coordinate at Xt and Xt-1; iii) average distance between adjacent Cα coordinates at Xt; iv) average rotation distance between a residue orientation at Xt and X0; v) average rotation distance between a residue orientation at Xt and Xt-1. B-C) While RFdiffusion is trained to generate samples over 200 timesteps, in many cases, trajectories can be shortened to improve computational efficiency. B) Larger steps can be taken between timesteps at inference. Decreasing the number of timesteps speeds up inference, and often does not decrease in silico success rates (left) (for example, on an NVIDIA A4000 GPU, 100 amino acid designs can be generated with 15 steps, in ~11s, with an in silico success rate of over 60%). When normalized for compute budget (center) it is often much more efficient to run more trajectories with fewer timesteps. This can be done without loss of diversity in samples (right). For harder problems (e.g. unconditional 300 amino acids), one must strike an intermediate number of total timesteps (e.g., T = 50) for optimal compute efficiency. Note that for all other analyses in the paper, 200 inference steps were used, in line with how RFdiffusion is trained. C) An alternative to taking larger steps is to stop trajectories early (possible because RFdiffusion predicts X0 at every timestep). In many cases, trajectories can be stopped at timestep 50–75 with little effect on the final in silico success rate of designs (left), and when normalized by compute budget (center), success rates per unit time are typically higher generating more designs with early-stopping. Again, this can be done without a significant loss in diversity (right).
A)对模拟的正向(噪声)和反向(去噪)轨迹的分析表明,Cα坐标的分布与残差方向的分布非常吻合,表明RF扩散已经按照预期学习了去噪过程的分布。从左到右:i) Cα 坐标在 X t 处与其在 X 0 中的位置之间的平均距离;ii) Cα 坐标在 X t 和 X t-1 处的平均距离;iii) 相邻 Cα 坐标在 X t 处的平均距离;iv) 残基取向在 X t 和 X 0 处的平均旋转距离;v) 残基取向在 X t 和 X t-1 处的平均旋转距离。B-C) 虽然 RFdiffusion 经过训练可以生成超过 200 个时间步长的样本,但在许多情况下,可以缩短轨迹以提高计算效率。B) 在推理时,可以在时间步长之间采取更大的步骤。减少时间步长的数量可以加快推理速度,并且通常不会降低计算机成功率(左图)(例如,在 NVIDIA A4000 GPU 上,可以通过 15 个步骤生成 100 个氨基酸设计,在 ~11 秒内,计算机成功率超过 60%)。当对计算预算(中心)进行归一化时,以更少的时间步长运行更多的轨迹通常效率更高。这可以在不损失样品多样性的情况下完成(右)。对于较难的问题(例如无条件的 300 个氨基酸),必须达到中间数量的总时间步长(例如,T = 50)以获得最佳计算效率。请注意,对于论文中的所有其他分析,使用了 200 个推理步骤,与 RFdiffusion 的训练方式一致。C) 采取更大步骤的另一种方法是尽早停止轨迹(可能,因为 RFdiffusion 在每个时间步长都预测 X 0 )。 在许多情况下,轨迹可以在时间步长 50-75 处停止,对设计的最终计算机成功率几乎没有影响(左图),当按计算预算(中心)进行归一化时,每单位时间的成功率通常更高,从而产生更多提前停止的设计。同样,这可以在不显着损失多样性的情况下完成(右)。
Extended Data Fig. 3 Unconditionally-generated designs are folded and thermostable.
扩展数据 图 3 无条件生成的设计是折叠的和热稳定的。
A) Four 200 amino acid and fourteen 300 amino acid proteins were tested for expression and stability. 9/18 designs expressed, with a major peak at the expected elution volume. Blue: 300 amino acid proteins; Purple: 200 amino acid proteins. B) Colored AF2 predictions overlaid on gray design models (left), circular dichroism spectra at 25 °C (blue) and 95 °C (pink) (middle) and circular dichroism melt curves (right) for all 9 designs passing expression thresholds. In all cases, proteins remain well folded even at 95 °C. Note that data on 300aa_3 and 300aa_8 are duplicated from Fig. 2f, reproduced here for clarity.
A) 测试了 4 个 200 个氨基酸和 14 个 300 个氨基酸蛋白的表达和稳定性。表达 9/18 设计,在预期洗脱体积处有一个主峰。蓝色:300个氨基酸蛋白质;紫色:200个氨基酸蛋白质。B) 覆盖在灰色设计模型上的彩色 AF2 预测(左)、25 °C(蓝色)和 95 °C(粉红色)(中)下的圆二色谱和所有 9 个通过表达阈值的设计的圆二色熔融曲线(右)。在所有情况下,即使在95°C下,蛋白质也能保持良好的折叠状态。 请注意,300aa_3和300aa_8的数据与图2f重复,为清楚起见,此处转载。
Extended Data Fig. 4 RFdiffusion can condition on fold information to generate specific, thermostable folds.
扩展数据 图 4 RF扩散可以调节褶皱信息以产生特定的、热稳定的褶皱。
A) 6WVS is a previously-described de novo designed TIM barrel (left). A fine-tuned RFdiffusion model can condition on 1D and 2D inputs representing this protein fold, specifically secondary structure (middle, bottom) and block-adjacency information (middle, top) (see Supplementary Methods 4.3.2). RFdiffusion then generates proteins that closely recapitulate this course-grained fold information (right). B) Outputs are diverse with respect to each other. With this coarse-grained fold specification, in silico successful designs are much more diverse (as quantified by pairwise TM-scores) compared to diversity generated through simply sampling many sequences for the original PDB backbone (6WVS). C) NTF2 folds are useful scaffolds for de novo enzyme design56, and can also be readily generated with fold-conditioning in RFdiffusion. Designs are diverse and closely recapitulated by AF2. D) In silico success rates are high with fold-conditioned diffusion. TIM barrels are generated with an AF2 in silico success rate of 42.5% (left bar, pink) with in silico success incorporating both AF2 metrics and a TM-score vs 6WVS > 0.5. NTF2 folds are generated with an AF2 in silico success rate of 54.1% (right bar, pink), with in silico success incorporating both AF2 metrics and a TM-score vs PDB: 1GY6 > 0.5. In silico success was further validated with ESMFold (blue bars), where a pLDDT > 80 was used as the confidence metric for success. Gray: RFdiffusion design, colors: AF2 prediction. E) 11 TIM barrel designs were purified alongside the 6WVS positive control. Ten of these express and elute predominantly as monomers (note that the designs are approximately 4kDa larger than 6WVS). F) Eight designs expressed sufficiently for analysis by circular dichroism. All designs are folded, with circular dichroism spectra consistent with the designed structure (middle), and similar to 6WVS. Designs were also all highly thermostable, with CD melt analyses demonstrating designs were folded even at 95 °C (right). Designs are shown in gray, with the AF2 predictions overlaid in colors (left). Note that data on 6WVS and TIM_barrel_6 are duplicated from Fig. 2g, reproduced here for clarity.
A) 6WVS 是前面描述的从头设计的 TIM 枪管(左)。微调的 RFdiffusion 模型可以调节代表该蛋白质折叠的 1D 和 2D 输入,特别是二级结构(中间、底部)和块邻接信息(中间、顶部)(参见补充方法 4.3.2)。然后,RFdiffusion产生蛋白质,这些蛋白质紧密地概括了这种过程粒度的折叠信息(右)。B) 产出彼此之间是多种多样的。有了这种粗粒度折叠规范,与通过简单地对原始PDB主链(6WVS)的许多序列进行采样而产生的多样性相比,计算机模拟成功的设计更加多样化(通过成对TM评分量化)。C) NTF2 折叠是酶从头设计的 56 有用支架,也可以很容易地通过 RFdifffusion 中的折叠调节生成。设计多种多样,并由 AF2 紧密概括。D) 在计算机模拟中,折叠条件扩散的成功率很高。TIM 桶的 AF2 计算机成功率为 42.5%(左栏,粉红色),计算机成功结合了 AF2 指标和 TM 分数与 6WVS > 0.5。NTF2 折叠的产生,AF2 计算机成功率为 54.1%(右栏,粉红色),计算机模拟成功结合了 AF2 指标和 TM 评分与 PDB:1GY6 > 0.5。ESMFold(蓝条)进一步验证了计算机成功,其中 pLDDT > 80 用作成功的置信度。灰色:RF扩散设计,颜色:AF2预测。E) 11 个 TIM 桶设计与 6WVS 阳性对照一起纯化。其中 10 种主要以单体形式表达和洗脱(请注意,这些设计比 6WVS 大约 4kDa)。F) 八种设计足以通过圆二色性进行分析。 所有设计均为折叠,圆二色光谱与设计结构(中)一致,与6WVS相似。设计也都具有高度的热稳定性,CD熔体分析表明,即使在95°C下,设计也能折叠(右图)。设计以灰色显示,AF2 预测以颜色覆盖(左)。请注意,6WVS和TIM_barrel_6的数据与图2g重复,为清楚起见,此处复制。
Extended Data Fig. 5 Symmetric oligomer design with RFdiffusion.
扩展数据 图 5 RFdiffusion 的对称低聚物设计。
A) Due to the (near-perfect - see Supplementary Methods 3.1) equivariance properties of RFdiffusion, X0 predictions from symmetric inputs are also symmetric, even at very early timepoints (and becoming increasingly symmetric through time; r.m.s.d. vs symmetrized: t = 200 1.20 Å; t = 150 0.40 Å; t = 50 0.06 Å; t = 0 0.02Å). Gray: symmetrized (top left) subunit; colors: RFdiffusion X0 prediction. B) In silico success rates for symmetric oligomer designs of various cyclic and dihedral symmetries. In silico success is defined here as the proportion of designs for which AF2 yields a prediction from a single sequence that has mean pLDDT > 80 and backbone r.m.s.d. over the oligomer between the design model and AF2 < 2Å. Note that 16 sequences per RFdiffusion design were sampled. C) Box plots of the distribution of backbone r.m.s.d.s between AF2 and the RFdiffusion design model with and without the use of external potentials during the trajectory. The external potentials used are the ‘inter-chain’ contact potential (pushing chains together), as well as the ‘intra-chain’ contact potential (making chains more globular). Using these potentials dramatically improves in silico success (Two-proportion z-test of in silico success rate: n = 100 designs per condition, z = 4.3, p = 1.9e-5). D) Designs are diverse with respect to the training dataset (the PDB). While the monomers (typically 60–100 AA) show reasonable alignment to the PDB (median 0.72), the whole oligomeric assemblies showed little resemblance to the PDB (median 0.50). E) Additional examples of design models (left) against AF2 predictions (right) for C3, C5, C12, and D4 symmetric designs (the symmetries not displayed in Fig. 3) with backbone r.m.s.d.s (Å) against their AF2 predictions of 0.82, 0.63, 0.79, and 0.78 with total amino acids 750, 900, 960, 640. F) Additional nsEM data for symmetric designs. The model is shown on the left and the 2D class averages on the right for each design. G) Two orthogonal side views of HE0537 by cryo-EM. Representative 2D class averages from the cryo-EM data are shown to the right of 2D projection images of the computational design model (lowpass filtered to 8 Å), which appear nearly identical to the experimental data. Scale bars shown (white) are 60 Å. Boxplot represents median ± IQR; tails: min/max excluding outliers (±1.5xIQR).
A) 由于 RFdiffusion 的(近乎完美 - 参见补充方法 3.1)等方差特性,来自对称输入的 X 0 预测也是对称的,即使在非常早期的时间点(并且随着时间的推移变得越来越对称;r.m.s.d. vs symmetrized:t = 200 1.20 Å;t = 150 0.40 Å;t = 50 0.06 Å;t = 0 0.02Å)。灰色:对称(左上)亚基;颜色:RFdiffusion X0 预测。B) 各种循环对称性和二面体对称性的对称低聚物设计的计算机成功率。在计算机模拟中,成功被定义为 AF2 从单个序列中产生预测的设计比例,该序列的平均 pLDDT > 80,并且设计模型和 AF2 < 2Å 之间的低聚物具有骨架 r.m.s.d.。请注意,每个 RFdiffusion 设计采样了 16 个序列。C) 在轨迹过程中使用外部电位和不使用外部电位的情况下,AF2 和 RFdiffusion 设计模型之间主干 r.m.s.d.s 分布的箱形图。使用的外部电位是“链间”接触电位(将链推到一起),以及“链内”接触电位(使链更呈球形)。利用这些潜力可以显著提高计算机模拟成功率(计算机模拟成功率的双比例 z 检验:每个条件 n = 100 个设计,z = 4.3,p = 1.9e-5)。D) 在训练数据集 (PDB) 方面,设计是多种多样的。虽然单体(通常为60-100 AA)与PDB的合理比对(中位数为0.72),但整个低聚体组装体与PDB的相似性不大(中位数为0.50)。E) 针对 C3、C5、C12 和 D4 对称设计(图 3 中未显示的对称性)的 AF2 预测(右)的设计模型(左)的其他示例,主干 r.m.s.d.s (Å) 与其 AF2 预测 0.82、0.63、0.79 和 0。78个,总氨基酸为750、900、960、640。F) 用于对称设计的附加 nsEM 数据。模型显示在左侧,右侧显示每个设计的 2D 类平均值。G) 冷冻电镜 HE0537 的两个正交侧视图。来自冷冻电镜数据的代表性 2D 类平均值显示在计算设计模型的 2D 投影图像(低通滤波至 8 Å)的右侧,这些图像看起来与实验数据几乎相同。显示的比例尺(白色)为 60 Å。箱线图表示 IQR ±中位数;尾部:最小值/最大值,不包括异常值 (±1.5xIQR)。
Extended Data Fig. 6 External potentials for generating pockets around substrate molecules.
扩展数据 图6 在底物分子周围产生口袋的外部电位。
A–D) Example in silico successful designs for enzyme classes 2–5 (ref. 57, see also Fig. 4). Native enzyme (PDB: 1CWY, 1DE3, 1P1X, 1SNZ); catalytic site (teal); RFdiffusion output (gray: model, colors: AF2 prediction). Metrics (AF2 vs design backbone r.m.s.d., AF2 vs design motif backbone r.m.s.d., AF2 vs design motif full-atom r.m.s.d., AF2 pAE): EC2: 0.93 Å, 0.50 Å, 1.29 Å, 3.51; EC3: 0.92 Å, 0.60 Å, 1.07 Å, 4.59; EC4: 0.93 Å, 0.80 Å, 1.03 Å, 4.41; EC5: 0.78 Å, 0.44 Å, 1.14 Å, 3.32. E–H) Implicit modeling of a substrate while scaffolding a retroaldolase active site triad [TYR1051-LYS1083-TYR1180] from PDB: 5AN7. E) The potential used to implicitly model the substrate, which has both a repulsive and attractive field (see Supplementary Methods 4.4). F) Left: Kernel densities demonstrate that without using the external potential (pink), designs often fall into two failure modes: (1) no pocket, and (2) clashes with the substrate. Right: clashes (substrate < 3 Å of the backbone) & pockets (no clash and > 16 Cα within 3–8 Å of substrate) with and without the potential. Two-proportion z-test: n = 71/51 +/− potential; clashes z = −2.05, p = 0.02, pocket z = −2.27, p = 0.01. Each datapoint represents a design already passing the stringent in silico success metrics (AF2 motif r.m.s.d. < 1 Å, AF2 backbone r.m.s.d. < 2 Å, AF2 pAE < 5). Note that the potential and clash definition pertain only to backbone Cα atoms, and do not currently include sidechain atoms. G) Designs close to the labeled local maxima of the kernel density estimate. Without the potential, the catalytic triad is predominantly (1) exposed on the surface with no residues available to provide substrate stabilization or (2) buried in the protein core, preventing substrate access. With the potential, the catalytic triad is predominantly (3), partially buried in a concave pocket with shape complementary to the substrate. Backbone atoms within 3 Å of the substrate are shown in red. H) A variety of diverse designs with pockets made using the potential, with no clashes between the substrate and the AF2-predicted backbone. The functional form and parameters used for the pocket potential are detailed in Supplementary Methods 4.4. In each case the substrate is superimposed on the AF2 prediction of the catalytic triad.
A-D)计算机成功设计酶类2-5的示例(参考文献 57 ,另见图4)。天然酶(PDB:1CWY、1DE3、1P1X、1SNZ);催化位点(蓝绿色);RFdiffusion 输出(灰色:模型,颜色:AF2 预测)。指标(AF2 与设计主干 r.m.s.d.、AF2 与设计基序主干 r.m.s.d.、AF2 与设计基序全原子 r.m.s.d.、AF2 pAE):EC2:0.93 Å、0.50 Å、1.29 Å、3.51;EC3: 0.92 å, 0.60 å, 1.07 å, 4.59;EC4: 0.93 å, 0.80 å, 1.03 å, 4.41;EC5: 0.78 å, 0.44 å, 1.14 å, 3.32.E-H) 来自 PDB:5AN7 的背醛缩酶活性位点三TYR1180 LYS1083 TYR1051联体支架时底物的隐式建模:5AN7。E) 用于隐式模拟基板的电位,该基板具有排斥场和吸引场(参见补充方法 4.4)。F) 左图:核密度表明,在不使用外部电位(粉红色)的情况下,设计通常陷入两种故障模式:(1) 无口袋,以及 (2) 与基板冲突。右图:有和没有电位的冲突(基板<3 Å的基板)和口袋(无冲突,在基板的3-8 Å内>16 Cα)。双比例 z 检验:n = 71/51 +/− 电位;冲突 z = −2.05,p = 0.02,口袋 z = −2.27,p = 0.01。每个数据点代表一个已经通过严格的计算机成功指标的设计(AF2 motif r.m.s.d. < 1 Å,AF2 backbone r.m.s.d. < 2 Å,AF2 pAE < 5)。请注意,势和冲突定义仅适用于主链 Cα 原子,目前不包括侧链原子。G) 接近核密度估计值的标记局部最大值的设计。如果没有电位,催化三联体主要 (1) 暴露在表面,没有残基可用于提供底物稳定或 (2) 埋在蛋白质核心中,阻止底物进入。 具有潜力的催化三联体主要是 (3),部分埋在凹形口袋中,形状与底物互补。基底 3 Å 范围内的主链原子以红色显示。H) 各种不同的设计,其口袋利用电位制成,基板和 AF2 预测的主链之间没有冲突。用于口袋电位的函数形式和参数详见补充方法 4.4。在每种情况下,底物都叠加在催化三元组的 AF2 预测上。
Extended Data Fig. 7 Additional Ni2+ binding C4 oligomers.
扩展数据 图7 额外的Ni 2+ 结合C4低聚物。
A) AF2 predictions of a subset of the experimentally verified Ni2+ binding oligomers, with corresponding isothermal titration calorimetry (ITC) binding isotherms for the wild-type (blue) and H52A mutant (pink) below. Note that these, with Fig. 5, encompass all of the experimentally validated outputs deriving from unique RFdiffusion backbones. Wild-type dissociation constants are displayed in each plot. We observe a mixture of endothermic (NiB2.10, NiB2.23, NiB2.15) and exothermic isotherms. For all cases displayed we observe no binding to the ion for H52A mutants, indicating the scaffolded histidine at position 52 is critical for ion binding. KD values in the isotherms indicate binding of the ion with the designed stoichiometry (1:4 Ni2+:protein). Note that each backbone depicted is from a unique RFdiffusion sampling trajectory, and that models and data for designs NiB2.15, NiB1.12, NiB1.20 and NiB1.17 from Fig. 5 are duplicated here for ease of viewing. B) Size exclusion chromatograms for elutions from the 44 purifications suggest the vast majority of designs are soluble and have the correct oligomeric state. C) Size exclusion chromatograms for 20 H52A mutants show that the mutants remain soluble and retain the intended oligomeric state. Note that only 18 of these 20 had wild-type sequences that definitively bound nickel. Note also that for ITC plots, points represent single measurements.
A) 对实验验证的 Ni 2+ 结合低聚物子集的 AF2 预测,以及下面野生型(蓝色)和 H52A 突变体(粉红色)的相应等温滴定量热法 (ITC) 结合等温线。请注意,这些输出与图 5 一样,包含了所有经过实验验证的输出,这些输出来自独特的 RFdiffusion 主干。野生型解离常数显示在每个图中。我们观察到吸热(NiB2.10、NiB2.23、NiB2.15)和放热等温线的混合物。对于显示的所有情况,我们观察到 H52A 突变体没有与离子结合,这表明 52 位的支架组氨酸对离子结合至关重要。等温线中的K D 值表示离子与设计的化学计量(1:4 Ni 2+ :p rotein)结合。请注意,所描绘的每个主干都来自唯一的RF扩散采样轨迹,并且图5中NiB2.15、NiB1.12、NiB1.20和NiB1.17设计的模型和数据在这里被复制,以便于查看。B) 44 种纯化洗脱液的尺寸排阻色谱图表明,绝大多数设计是可溶的,并且具有正确的低聚状态。C) 20 个 H52A 突变体的尺寸排阻色谱图显示突变体保持可溶性并保留预期的低聚体状态。请注意,这 20 个序列中只有 18 个具有明确结合镍的野生型序列。另请注意,对于 ITC 图,点表示单个测量值。
Extended Data Fig. 8 Targeted unconditional and fold-conditioned protein binder design.
扩展数据 图8 靶向无条件和折叠条件蛋白质结合剂设计。
A-B) The ability to specify where on a target a designed binder should bind is crucial. Specific “hotspot” residues can be input to a fine-tuned RFdiffusion model, and with these inputs, binders almost universally target the correct site. A) IL-7Rα (PDB: 3DI3) has two patches that are optimal for binding, denoted Site 1 and Site 2 here. For each site, 100 designs were generated (without fold-specification). B) Without guidance, designs typically target Site 1 (left bar, gray), with contact defined as Cα-Cα distance between binder and hotspot reside < 10 Å. Specifying Site 1 hotspot residues increases further the efficiency with which Site 1 is targeted (left bar, pink). In contrast, specifying the Site 2 hotspot residues can completely redirect RFdiffusion, allowing it to efficiently target this site (right bar, pink). C-D) As well as conditioning on hotspot residue information, a fine-tuned RFdiffusion model can also condition on input fold information (secondary structure and block-adjacency information - see Supplementary Methods 4.5). This effectively allows the specification of a (for instance, particularly compatible) fold that the binder should adopt. C) Two examples showing binders can be specified to adopt either a ferredoxin fold (left) or a particular helical bundle fold (right). D) Quantification of the efficiency of fold-conditioning. Secondary structure inputs were accurately respected (top, pink). Note that in this design target and target site, RFdiffusion without fold-specification made generally helical designs (right, gray bar). Block-adjacency inputs were also respected for both input folds (bottom, pink). E) Reducing the noise added at each step of inference improves the quality of binders designed with RFdiffusion, both with and without fold-conditioning. As an example, the distribution of AF2 interaction pAEs (known to indicate binding when pAE < 1026) is shown for binders designed to PD-L1. In both cases, the proportion of designs with interaction pAE < 10 is high (blue curve), and improved when the noise is scaled by a factor 0.5 (pink curve) or 0 (yellow curve). F) Full in silico success rates for the protein binders designed to five targets. In each case, the best fold-conditioned results are shown (i.e. from the most target-compatible input fold), and the success rates at each noise scale are separated. In line with current best practice26, we tested using Rosetta FastRelax58 before designing the sequence with ProteinMPNN, but found that this did not systematically improve designs. In silico success is defined in line with current best practice26: AF2 pLDDT of the monomer > 80, AF2 interaction pAE < 10, AF2 r.m.s.d. monomer vs design < 1 Å. G) Experimentally-validated de novo protein binders were identified for all five of the targets. Designs that bound at 10 μM during single point BLI screening with a response equal to or greater than 50% of the positive control were considered binders. Concentration is denoted by hue for designs that were screened at concentrations less than 10 μM and thus may be false negatives.
A-B) 指定设计粘合剂应在目标上结合的位置的能力至关重要。特定的“热点”残基可以输入到微调的RFdiffusion模型中,通过这些输入,粘合剂几乎普遍靶向正确的位点。A) IL-7Rα (PDB: 3DI3) 有两个最适合结合的贴片,此处表示为 Site 1 和 Site 2。对于每个站点,生成了 100 个设计(没有折叠规范)。B) 在没有指导的情况下,设计通常以站点 1(左条,灰色)为目标,接触定义为粘合剂和热点之间的 Cα-Cα 距离< 10 Å。 指定站点 1 热点残留物可进一步提高靶向站点 1 的效率(左条,粉红色)。相比之下,指定站点 2 热点残留物可以完全重定向 RFdiffusion,使其能够有效地定位该站点(右栏,粉红色)。C-D) 除了对热点残基信息进行调节外,微调的 RFdiffusion 模型还可以对输入折叠信息(二级结构和块邻接信息 - 参见补充方法 4.5)进行调节。这有效地允许对粘合剂应采用的(例如,特别兼容的)折叠进行规范。C) 可以指定两个显示粘合剂的示例,以采用铁氧还蛋白折叠(左)或特定的螺旋束折叠(右)。D) 折叠调节效率的量化。二级结构输入得到准确遵守(顶部,粉红色)。请注意,在此设计目标和目标站点中,没有折叠规范的 RFdiffusion 通常采用螺旋设计(右图,灰色条)。两个输入折叠(底部,粉红色)也都考虑了块邻接输入。 E) 减少推理每一步添加的噪声可以提高使用 RFdiffusion 设计的粘合剂的质量,无论是否具有折叠调节功能。例如,对于设计用于 PD-L1 的结合剂,显示了 AF2 相互作用 pAE(已知在 pAE < 10 26 时指示结合)的分布。在这两种情况下,具有交互作用 pAE 的设计比例< 10 表示高(蓝色曲线),当噪声缩放系数 0.5(粉红色曲线)或 0(黄色曲线)时,噪声会得到改善。 F) 设计用于五个靶标的蛋白质结合剂的全计算机成功率。 在每种情况下,都会显示最佳折叠条件结果(即来自最兼容目标的输入折叠),并且每个噪声标度下的成功率都是分开的。 根据当前的最佳实践 26 ,我们使用 Rosetta FastRelax 58 进行了测试之前用 ProteinMPNN 设计序列,但发现这并没有系统地改进设计。计算机模拟成功与当前最佳实践一致 26 :单体的AF2 pLDDT>80,AF2相互作用pAE<10,AF2 r.m.s.d.单体与设计<1 Å。 G) 针对所有五个靶标鉴定了经过实验验证的从头蛋白质结合剂。在单点 BLI 筛选期间以 10 μM 结合且响应等于或大于阳性对照 50% 的设计被视为结合剂。对于在浓度低于 10 μM 下筛选的设计,浓度用色调表示,因此可能是假阴性。
Extended Data Fig. 9 Cryo-electron microscopy structure determination of designed Influenza HA binder.
扩展数据 图9 冷冻电子显微镜测定设计的流感HA粘合剂。
A) Representative raw micrograph showing ideal particle distribution and contrast. B) 2D Class averages of Influenza H1+HA_20 binder with clearly defined secondary structure elements and a full-sampling of particle view angles (scale bar = 10 nm). C) Cryo-EM local resolution map calculated using an FSC value of 0.143 viewed along two different angles. Local resolution estimates range from ~2.3 Å at the core of H1 to ~3.4 Å along the periphery of the N-terminal helix of the HA_20 binder. D) Cryo-EM structure of the full H1+HA_20 binder complex (purple: HA_20; yellow: H1; teal: glycans). E) Global resolution estimation plot. F) Orientational distribution plot demonstrating complete angular sampling. G) 3D ab initio (left) and 3D heterogenous refinement (right - unsharpened) outputs, performed in the absence of applied symmetry, and showing clear density of the HA_20 binder bound to all three stem epitopes of the Iowa43 HA glycoprotein trimer, in all maps. H) The designed binder has topological similarity to 5VLI, a protein in the PDB, but binds with very different interface contacts.
A) 具有代表性的原始显微照片,显示理想的颗粒分布和对比度。B) 具有明确定义的二级结构元素和颗粒视角全采样(比例尺 = 10 nm)的 H1+HA_20 流感粘合剂的 2D 类平均值。C) 冷冻电镜局部分辨率图,使用 FSC 值 0.143 沿两个不同角度观察计算。局部分辨率估计范围从H1核心的~2.3 Å到HA_20粘合剂N端螺旋外围的~3.4 Å。D)完整H1+HA_20结合剂复合物的冷冻电镜结构(紫色:HA_20;黄色:H1;蓝绿色:聚糖)。E) 全局分辨率估计图。F) 显示完整角度采样的取向分布图。G) 3D 从头开始(左)和 3D 异质细化(右 - 未锐化)输出,在没有应用对称性的情况下进行,并在所有图谱中显示与 Iowa43 HA 糖蛋白三聚体的所有三个茎表位结合的 HA_20 结合剂的清晰密度。H) 设计的结合剂与 PDB 中的一种蛋白质 5VLI 具有拓扑相似性,但与非常不同的界面接触结合。
扩展数据表1 冷冻电镜数据收集、改进和验证统计
Supplementary information
补充资料
Supplementary Information
补充资料
The supplementary information file is a single PDF that contains text, figures and tables that aim to help the reader understand the theoretical underpinnings of RFdiffusion, its implementation and its application to the design challenges posed in the paper. Descriptions of in silico and experimental methods can be found within.
补充信息文件是一个单独的 PDF,其中包含文本、图形和表格,旨在帮助读者了解 RFdiffusion 的理论基础、其实现及其在论文中提出的设计挑战中的应用。计算机模拟和实验方法的描述可以在其中找到。
Rights and permissions 权利和权限
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
开放获取 本文根据知识共享署名 4.0 国际许可获得许可,该许可允许以任何媒介或格式使用、共享、改编、分发和复制,只要您适当注明原作者和来源,提供知识共享许可的链接,并说明是否进行了更改。本文中的图像或其他第三方材料包含在文章的知识共享许可中,除非在材料的鸣谢中另有说明。如果文章的知识共享许可中未包含材料,并且您的预期用途不受法律法规允许或超出允许的用途,则需要直接获得版权所有者的许可。要查看此许可证的副本,请访问 http://creativecommons.org/licenses/by/4.0/。
About this article
Cite this article
Watson, J.L., Juergens, D., Bennett, N.R. et al. De novo design of protein structure and function with RFdiffusion. Nature 620, 1089–1100 (2023). https://doi.org/10.1038/s41586-023-06415-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-023-06415-8
This article is cited by
本文被引用
-
Enzyme engineering for functional lipids synthesis: recent advance and perspective
功能性脂质合成的酶工程研究进展与展望Bioresources and Bioprocessing (2024)
生物资源和生物加工 (2024) -
State-specific protein–ligand complex structure prediction with a multiscale deep generative model
基于多尺度深度生成模型的状态特异性蛋白质-配体复合物结构预测Nature Machine Intelligence (2024)
自然机器智能 (2024) -
Piccadilly full of people and other foul things
皮卡迪利到处都是人和其他肮脏的东西EMBO Reports (2024) EMBO 报告 (2024)
-
Targeted genome-modification tools and their advanced applications in crop breeding
靶向基因组修饰工具及其在作物育种中的先进应用Nature Reviews Genetics (2024)
自然评论遗传学 (2024) -
Biosensor and machine learning-aided engineering of an amaryllidaceae enzyme
amaryllidaceae酶的生物传感器和机器学习辅助工程Nature Communications (2024)
自然通讯 (2024)
Comments 评论
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
提交评论即表示您同意遵守我们的条款和社区准则。如果您发现有辱骂性内容或不符合我们的条款或指南的内容,请将其标记为不当内容。